Abstract
A group of EGFR inhibitors derived from thieno[2,3-d]pyrimidine nucleus was designed, synthesised, and examined as anti-proliferative lead compounds. MCF-7 and A549 cell lines were inhibited by 5b, the most active member. It had inhibitory partialities of 37.19 and 204.10 nM against EGFRWT and EGFRT790M, respectively. Compound 5b was 2.5 times safer against the WI-38 normal cell lines than erlotinib. Also, it demonstrated considerable potentialities for both early and late apoptosis induction in A549. Simultaneously, 5b arrested A549’s growth at G1 and G2/M phases. Harmoniously, 5b upregulated the BAX and downregulated the Bcl-2 genes by 3-fold and increased the BAX/Bcl-2 ratio by 8.3-fold comparing the untreated A549 cells. Molecular docking against EGFRWT and EGFRT790M indicated the correct binding modes. Furthermore, MD simulations confirmed the precise binding of 5b against the EGFR protein over 100 ns. Finally, various computational ADMET studies were carried out and indicated high degrees of drug-likeness and safety.
Introduction
Despite the tremendous efforts to confront it, cancer remains one of the biggest public health problems facing humanityCitation1. According to the WHO in 2020, about 10 million deaths, or nearly one in six deaths, were caused by cancer, making it the top cause of death globallyCitation2. Therefore, cancer is considered a great problem for the economy of countries as well as individualsCitation3,Citation4. Oncogenic mutations, multi-drug resistance, and activation of compensatory mechanisms are examples of how the complication of cancer pathology expresses itselfCitation5–7. Therefore, it is imperative to discover anticancer candidates that are more effective and less toxic, depending on the different biological and molecular features of cancer pathogenesis.
The protein kinases family which comprises huge types of vital enzymes and control different cellular processes including the proliferation and metastasis of human tumour cellsCitation8. Epidermal growth factor receptor (EGFR) is an important member of this familyCitation9 that has a crucial role in the growth of tumour cellsCitation10. Overexpression of EGFR is an effective prognostic factor in many types of tumours. Accordingly, hindering its molecular role exerts substantial therapeutic impactsCitation11. Thus, EGFR drew attention as an influential factor in the fight against cancerCitation12–14.
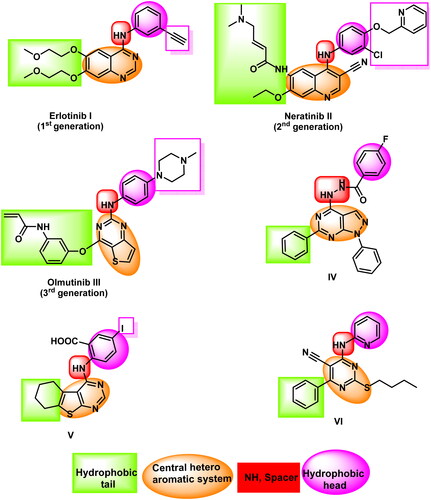
Erlotinib, I, is a member of the first generation of FDA-approved EGFR inhibitors exhibiting an excellent inhibitory effect against non-small-cell lung cancerCitation15. The appearance of EGFR mutation-related resistance led scientists to discover the second generation of FDA-approved EGFR inhibitors such as neratinib IICitation16. Unfortunately, the second generation inhibitors showed significant induced toxicityCitation17 leading to the invention of the third generation of EGFR inhibitors such as olmutinib IIICitation18,Citation19 (.
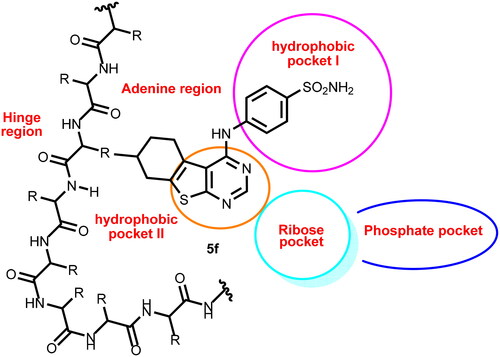
EGFR inhibitors share specific pharmacophoric features which are vital for the correct binding inside the ATP binding site of the EGFR protein. These essential features are represented in and including a flat hetero aromatic ringCitation20, a terminal hydrophobic head, an imino (NH, spacer) groupCitation21, and a hydrophobic tailCitation22,Citation23. The mentioned groups are essential to occupy the adenine binding pocket, the hydrophobic region I, the space between the adenine binding region and the hydrophobic region I, and the hydrophobic region II, respectively.
Figure 2. Compound 5f as a representative example of the synthesised compounds occupying the ATP active cavity of EGFR (schematic presentation).
Thieno[2,3-d]pyrimidine is an important scaffold as it is a building block in many biologically active agents as anticancerCitation24,Citation25, anti-viralCitation26, anti-oxidantCitation27, anti-inflammatoryCitation28,Citation29, and antimicrobialCitation30. In addition, thieno[2,3-d]pyrimidine showed good contributions in tyrosine kinase inhibitorsCitation31. Particularly, it was a cornerstone of many EGFR inhibitorsCitation32.
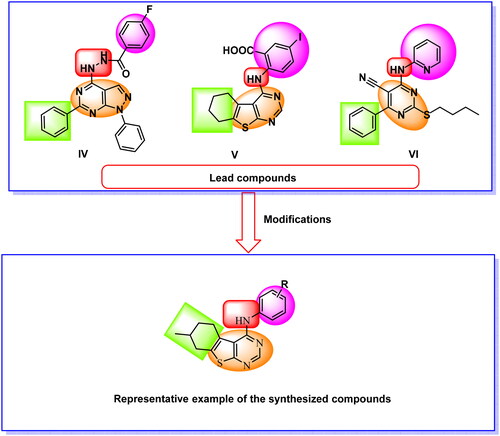
Our research team developed several 1H-pyrazolo[3,4-d]pyrimidine derivatives in 2018. The IC50 results for compound IV against the wild-type EGFRWT and mutant EGFRT790M were very promisingCitation33. In order to create an EGFR and HER2 tyrosine kinase inhibitor in 2019, we synthesised many thieno[2,3-d]pyrimidine derivatives. Promising action was seen in compound VCitation34. As EGFR inhibitors, we created a fresh batch of pyrimidine-5-carbonitrile derivatives in 2020. Compound VI exhibited strong inhibitory effects on the EGFRWT and EGFRT790MCitation35 (.
In this field, we used three biological isosteres (1H-pyrazolo[3,4-d]pyrimidine, thieno[2,3-d]pyrimidine, and pyrimidine-5-carbonitrile) to occupy the adenine binding pocket of EGFR. It was found that thieno[2,3-d]pyrimidine derivatives have good activities. Accordingly, we decided to modify these derivatives hoping to reach more effective anticancer agents targeting EGFR (.
The rationale of molecular design
In the current work, the previously reported anti-EGFR compounds (IV, V, and VI) were used as lead compounds to design new EGFR inhibitors. We focussed on the thieno[2,3-d]pyrimidine nucleus to reach the new members. Such moiety was used as a hetero aromatic system to occupy the adenine binding pocket of EGFR. This moisty involves three hydrogen bond acceptors (two nitrogen atoms and a sulphur atom). This may facilitate the hydrogen bonding at the adenine binding pocket of EGFR. In addition, the fused thieno[2,3-d]pyrimidine is large enough to fill the slightly large adenine binding pocketCitation36. Furthermore, the thieno[2,3-d]pyrimidine moiety is the main nucleus of the third-generation FDA-approved EGFR inhibitor (olmutinib III) and reported EGFR inhibitor (compound V). Also, the thieno[2,3-d]pyrimidine is considered as an isostere moiety for the 1H-pyrazolo[3,4-d]pyrimidine moiety of compound IV.
The position of NH liker was kept as it is. For the hydrophobic head, we used different structures as (i) substituted phenyl moieties (compounds 5a–f), (ii) benzyl moiety (compound 6), (iii) bulky hetero-aromatic structure (compound 8), and (iv) hydrophilic moieties as thiourea group (compounds 7a) and amino group (compound 7b).
Regarding the hydrophobic tail, we used the 4-methylcyclohex-1-ene moiety in all the synthesised compounds. The 4-methylcyclohex-1-ene moiety was used to increase the hydrophobic character of the hydrophobic tail. This may increase the binding power of the new compounds in the hydrophobic pocket II (.
The synthesised compounds were evaluated for it anti-proliferative activities against two tumour cell lines. The most active members were further evaluated for their anti-EGFR activity. Deep biological investigations such as cell cycle and apoptosis analyses were conducted for the most active member. Docking studies were carried out against wild and mutant EGFR to assess the binding mode of the synthesised compounds.
Results and discussion
Chemistry
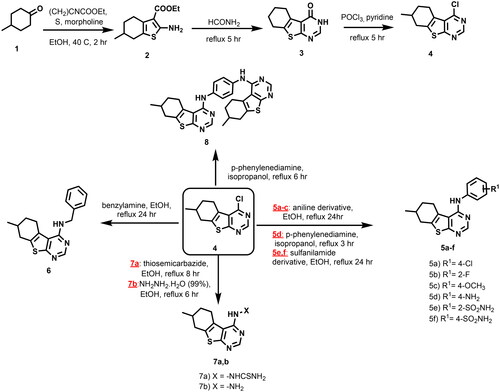
The target compounds 5a–f, 6, 7a,b, and 8 were synthesised as illustrated in Scheme 1. 4-Chloro-7-methyl-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidine 4Citation37, the key intermediate, was prepared via Gewald reaction of 4-methyl cyclohexanone 1 to give the amino ester derivative 2Citation38, which underwent cyclisation into the corresponding tricyclic 4-pyrimidione derivative 3 using formamide (99.5%)Citation37. Chlorination of the latter with POCl3 afforded the key intermediate tricyclic 4-chloropyrimidine 4Citation37. Thereafter, 4-chloropyrimidine 4 was allowed to undergo several nucleophilic aromatic substitution reactions with aniline derivatives in acetonitrile or absolute ethanol to afford the target compounds 5a–cCitation39. Also, the reaction of the key intermediate 4 with p-phenylenediamine in isopropanolCitation40 afforded compound 5d. The product 5d was isolated via pouring the reaction mixture on ice. Furthermore, heating of compound 4 with benzene sulphonamide derivatives, benzyl amine, thiosemicarbazide and hydrazine hydrate (99.9%) in absolute ethanol resulted in the formation of the desired compounds 5e–f, 6, and 7a, b respectively. Finally, heating of compound 4 under reflux with p-phenylenediamine in isopropanol with stirring for 6 h afforded compound 8. The formed dimer, unexpected product 8, was obtained via filtration of the reaction mixture while hot.
Biology
In vitro evaluation of anti-proliferative activity
Using the conventional MTT methodCitation41–43, all the synthesised compounds were tested for their in vitro anti-proliferative effects against a panel of two human carcinoma cell lines, the breast (MCF-7) and the lung (A549), both of which have overexpressed EGFRCitation44–46. The data in demonstrated that different synthetic candidates had anti-proliferative effects on the cell lines under examination. Although compounds were generally less effective against the two tested cell lines than erlotinib, compounds 5b and 5f were the noticeably active members against A549 cells (IC50 values of 17.79 and 17.46 µM) and against MCF-7 cells (IC50 values of 22.66 and 21.40 µM) comparing erlotinib (IC50 values of 4.18 and 14.27 µM), respectively. On the other side, the rest of the candidates, except compound 8, were moderately active expressing IC50 values ranging from 18.39 to 35.68 µM.
Table 1. In vitro anti-proliferative and EGFR inhibitory activities of the tested compounds.
Structure-activity relationships
Comparing the cytotoxicity of compounds 5a–f (incorporating substituted phenyl moieties), compound 6 (incorporating a benzyl moiety), and compound 8 (incorporating a bulky hetero-aromatic structure), revealed that the substituted phenyl moieties are more advantageous than the benzyl moiety, and the latter is more preferred than the bulky hetero-aromatic structure.
Regarding the substituted phenyl moieties (compounds 5a–f), it was found that the substitution with electron-withdrawing groups (compounds 5a, 5b, 5e, and 5f) is more advantageous than the substitution with electron-donating ones (compounds 5c and 5d). For the substitution with electron-withdrawing groups at the 4-position of the phenyl ring (compounds 5a and 5f), it was found the SO2NH2 group is more active than the chloro atom. For the substitution with electron-withdrawing groups at the 2-position of the phenyl ring (compounds 5b and 5e), it was found the fluoro atom is more active than the SO2NH2 group. Regarding the substitution with electron-donating groups at the 4-position of the phenyl ring (compounds 5c and 5d), it was found the OCH3 group is more active than the NH2 group. Finally, comparing the activities of compounds 7a (incorporating thiourea moiety) and 7b (incorporating amino group), it was found that the amino group is more advantageous than the thiourea moiety.
Toxicity of compound 5b towards normal human cells
The unintended toxicity caused to healthy cells by the anticancer drugs is one of the key serious side effects of chemotherapy. Also, it’s essential to examine if the obtained anti-proliferative activities of 5b originated from its anticancer potentiality or general toxicity. Accordingly, normal human lung fibroblasts, WI-38, cell lines were utilised to evaluate the safety of 5b. ’s findings regarding 5b’s in vitro anti-proliferative activities demonstrate that it appeared to be non-toxic to WI-38 with an IC50 of 70.13 µM compared to that of erlotinib (28.48 µM).
Evaluation of kinases activity
For the active candidate, 5b, against EGFRWT and EGFRT790M, a homogeneous time-resolved fluorescence (HTRF) assayCitation47 was used to assess its inhibitory activities. As a reference standard, erlotinib was used. With IC50 values of 37.19 and 204.10 nM against EGFRWT and mutant EGFRT790M, compound 5b had comparable inhibitory actions to those of erlotinib (IC50 = 5.9 and 212.2 nM, respectively) ().
Cell cycle analysis
Eukaryotic cells can replicate themselves through the successive cell cycle. Any anti-proliferative drug affects the targeted cells and stops their growth at one or more checkpoints (phases) of growth. If cellular damage persists, checkpoint signalling may trigger several systems that start apoptosis. Finding such a likely stage of cell arrest is essential for the drug discovery processCitation48.
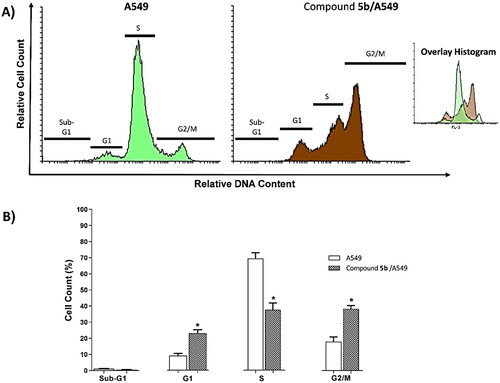
In this work, A549 cells were subjected to 5b at a concentration of 17.79 µM for 72h. Investigating the changes that occurred in the cell cycle, it was noticed that 5b induced a significant decrease in cells’ population at the S phase (37.66%) and at the Sub-G1 phase (0.54%), comparing to the untreated A549 cells (71.62% and 1.14%, respectively). Simultaneously, 5b exerted a substantial increase in the A549 population at the G1 phase (23.19%), and at the G2/M phase (38.61%), comparing to the untreated A549 cells (9.12% and 18.12%, respectively) (. This indicated that compound 5b can arrest the A549 cells at both G1 and G2/M phases ().
Table 2. Effect of 5b on cell cycle progression in A549 cells after 72 h treatment.
Apoptosis analysis
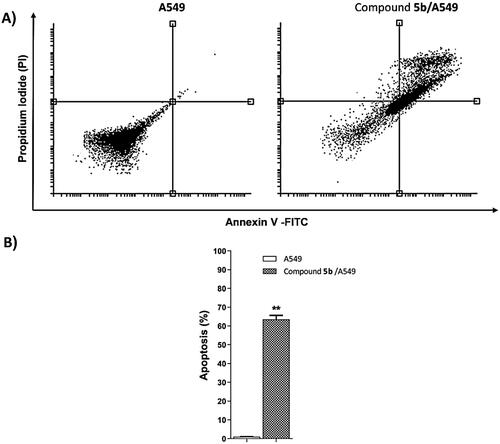
The apoptotic effect of 5b, 17.79 µM, on A549 cells was evaluated using the Annexin-V/propidium iodide staining (Annexin-V FTIC) test. As shown in and , compound 5b significantly increased and induced apoptosis (early and late) in the A549 cells to be 17.84% and 45.71%, respectively comparing the untreated A549 cells (0.07% and 0.98%, respectively.
Table 3. Effect of compound 5b on stages of the cell death process in A549 cells after 72 h treatment.
Effects on the mitochondrial apoptosis pathway
BAX and Bcl-2 are two primary regulating genes controlling mitochondrial apoptosis. During the apoptotic cascade, these mediators serve opposing functions. In contrast with Bcl-2 (anti-apoptotic), BAX exhibits a pro-apoptotic activity [43]. Therefore, the BAX/Bcl-2 ratio is a key factor to understand mitochondrial apoptosis [44]. In this test, A549 cells were treated with compound 5b at a concentration of 17.79 µM for 72h. The investigation of BAX and Bcl-2 genes expression by quantitative RT-PCR ( and ) showed the significant effects of 5b (17.79 µM) against the A549 cells. Interestingly, 5b upregulated the BAX and downregulated the Bcl-2 genes by 3-fold each. Simultaneously, 5b increased the BAX/Bcl-2 ratio by 8.3 times comparing the untreated A549 cells.
Figure 6. Gene expression analysis of BAX and Bcl-2 the expression levels after treatment of A549 with compound 5b for 72h.
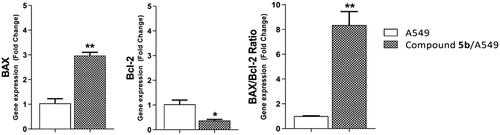
Table 4. Effect of 5b on levels of BAX and Bcl-2 genes expression in A549 cells treated for 72 h.
In silico studies
Docking studies
The docking studies were carried out against two forms of EGFR (Wild-type (EGFRWT, PDB: 4HJO)Citation49 and mutant type (EGFRT790M, PDB: 3W2O)Citation50) to assess the binding efficiency of the synthesised compounds against wild and mutant types of the EGFR protein. The co-crystallised ligands (erlotinib and TAK-285) were used as reference molecules. MOE 2019 software was used in these studies. The binding scores of the synthesised compounds and the co-crystallised ligands were summarised in .
Table 5. The docking binding free energies of the synthesised compounds against EGFRWT and EGFRT790M.
The binding pattern of erlotinib was similar to the reported resultsCitation35. It showed a binding score of −23.94 kcal/mol. Two hydrogen bonds (HBs) and seven hydrophobic interactions (HIs) stabilised the molecule in the active site. Each function group occupied a particular pocket in the active site as appeared in .
Figure 7. Erlotinib docked into the active site of EGFRWT forming two HBs with Met769 and Cys773 and seven HIs with Lys721, Val702, Ala719, Leu820, and Leu694.
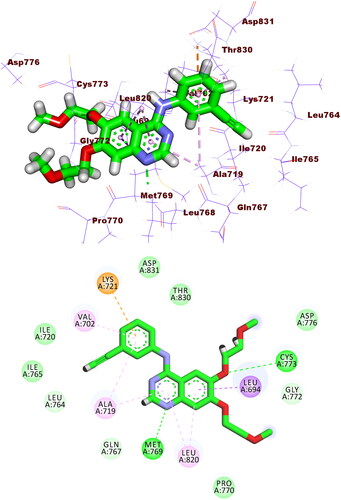
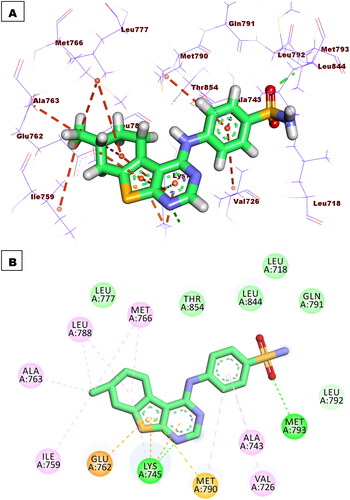
Compound 5b exhibited a similar binding pattern to erlotinib. It showed a binding score of −17.22 kcal/mol. It formed one HBs with Met769. In addition, it formed twelve HIs with Lys721, Val702, Ala719, Leu820, Cys773, and Leu694. The different moieties of compound 5b occupied the active pockets of EGFR. The thieno[2,3-d]pyrimidine, fluorobenzene, and 4-methylcyclohex-1-ene moieties occupied the adenine pocket, the hydrophobic pocket I, and the hydrophobic pocket II, respectively (.
Figure 8. Compound 5b docked into the active site of EGFRWT forming 1 HB with Met769 and 12 HIs with Lys721, Val702, Ala719, Leu820, Cys773, and Leu694.
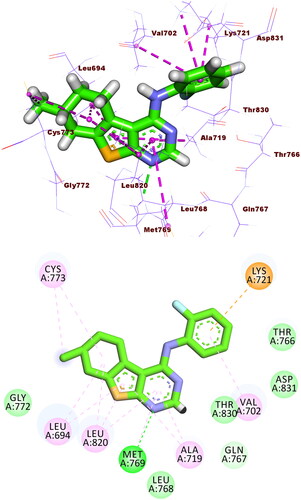
Compound 5f exhibited showed a binding score of −17.49 kcal/mol with a similar binding mode to erlotinib. It formed an HB with Met769 and ten HIs with Val702, Ala719, Leu820, Cys773, and Leu694. The thieno[2,3-d]pyrimidine, benzenesulphonamide, and 4-methylcyclohex-1-ene moieties occupied the adenine pocket, the hydrophobic pocket I, and the hydrophobic pocket II, respectively (.
Figure 9. Compound 5f docked into the active site of EGFRWT, forming 1 HB with Met769, and 10 HIs with Val702, Ala719, Leu820, Cys773, and Leu694.
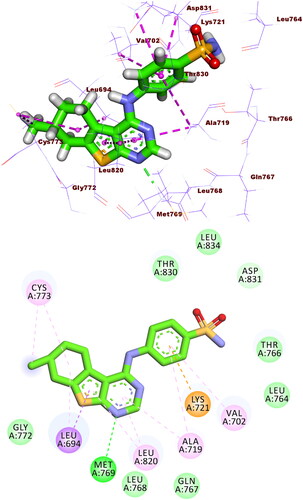
The binding modes of the tested compounds against EGFRT790M were similar to that of the co-crystallised ligand (TAK-285). The latter showed a binding score of −21.55 kcal/mol. It formed three HBs with Met793, Ser720, and Lys745. In addition, it formed fourteen HIs with Lys745, Glu762, Leu788, Ile759, Leu844, Ala743, Val726, Met790, and Ala743. The pyrrolo[3,2-d]pyrimidine, 3-(trifluoromethyl)phenoxy, N-ethyl-3-hydroxy-3-methylbutanamide moieties were buried in the adenine pocket, hydrophobic pocket I, and hydrophobic region II, respectively (.
Figure 10. Co-crystallised ligand (TAK-285) docked into the active site of EGFRT790M forming formed 3 HBs with Met793, Ser720, and Lys745 and 14 HIs with Lys745, Glu762, Leu788, Ile759, Leu844, Ala743, Val726, Met790, and Ala743.
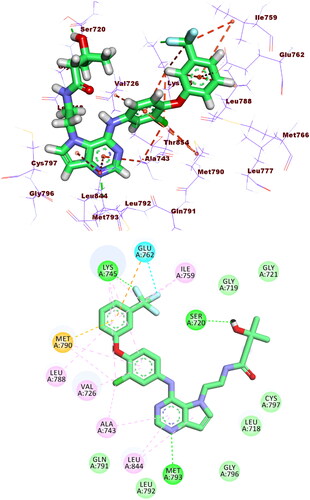
Compound 5f exhibited showed a binding score of −17.37 kcal/mol with a similar binding mode to TAK-285. It formed two HBs with Met793 and Lys745. In addition, it formed nine HBs with Met766, Leu788, Ile759, Val726, Met790, Ala763, and Ala743. Furthermore, it formed three electrostatic attractions with Met790, Lys745, and Glu762. The thieno[2,3-d]pyrimidine, benzenesulphonamide, and 4-methylcyclohex-1-ene moieties occupied the adenine pocket, the hydrophobic pocket I, and the hydrophobic pocket II, respectively (.
Molecular dynamic simulations
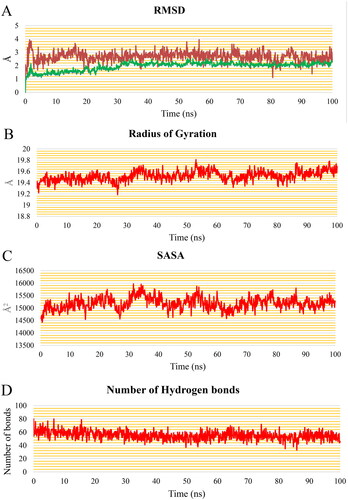
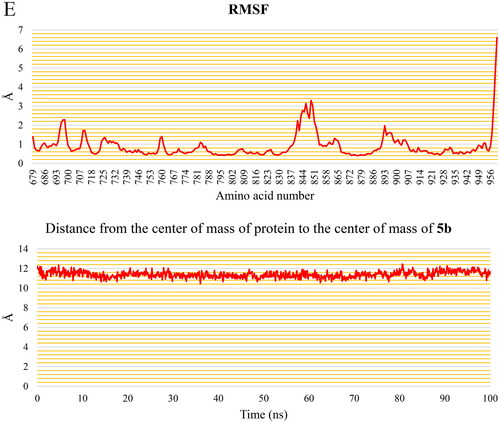
The EGFR protein shows consistent behaviour with a steady fluctuation of the 5b molecule, according to the analysis done for the EGFR-5b complex throughout the production run. (blue and green curves) shows that the EGFR and the EGFR-5b complex RMSD plots stabilise after 32 ns, with an average value of 2.11 Å for both of them for the rest of the trajectory. Furthermore, the RMSD of the 5b demonstrates a consistent trend around 2.69 Å throughout the trajectory. The RoG () exhibits a very little rising trend, reaching 19.7 Å at the end of the simulation with a change of only 0.4 Å from the initial frame. The SASA () shows a consistent average of 15 188 Å2. depicts a decrease in the number of HBs throughout the first 32 ns until it stabilises at an average of 53 HBs which indicates the stability of the protein conformation. Except for Ser696:Phe699, Ala840:Lys851, and the free C-terminal, which reach 2.2 Å, 3.3 Å, and 6.6 Å, respectively, the variation of the amino acids represented in the RMSF plot () is relatively low (less than 2 Å). The 5b has a constant separation of 11.3 Å between its centre of mass and the protein’s centre of mass ().
Figure 12. (A) RMSD values from the trajectory for the EGFR protein (blue curve), 5b (red curve), and EGFR & 5b complex (green curve), (B) radius of gyration, (C) SASA, (D) change in the number of HBs, (E) RMSF, and (F) distance from the centre of mass of 5b and EGFR protein.
MM-GBSA analysis
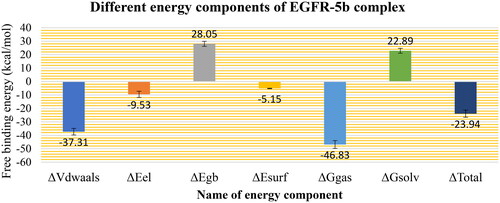
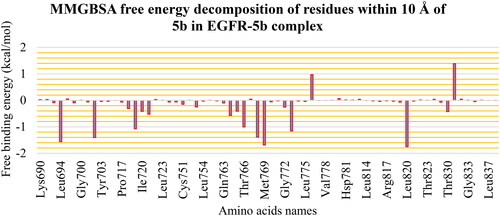
The different components that contribute to the MM-GBSA binding free energy analysis are depicted in . The overall binding of the 5b is −23.94 Kcal/Mol on average. The van der Waals energy has the most beneficial contribution with an average value of −37.31 Kcal/Mol, followed by the electrostatic interaction energy with an average value of −9.53 Kcal/Mol. Furthermore, we performed decomposition analysis () to determine which amino acids within 1 nm of the 5b molecule contribute to the interaction with a value less than −1 kcal/mol. These amino acids are Leu694 (−1.56 Kcal/Mol), Val702 (−1.4 Kcal/Mol), Ala719 (−1.07 Kcal/Mol), Thr766 (−1.0 Kcal/Mol), Leu768 (−1.39 Kcal/Mol), Met769 (−1.68 Kcal/Mol), Cys773 (−1.15 Kcal/Mol), and Leu820 (−1.75 Kcal/Mol). Moreover, two amino acids have positive free energy contributions to the binding (Asp776 (+0.97 Kcal/Mol) and Asp831 (+1.39 Kcal/Mol)).
PLIP analysis
The trajectory of the EGFR-5b complex was clustered, and then a representative frame for each cluster was generated. The elbow method was used to determine the number of clusters, and four clusters were generated. The PLIP website was used to detect the number and type of interactions between the 5b and the EGFR protein for each cluster representative. displays the number and types of interactions retrieved from the PLIP website. Two types of interactions have been reported from the PLIP webserver with a large difference in their numbers. Only one amino acid (Met769) is forming H-bond in all representatives. On the other hand, there are 20 HIs detected with Leu694, Thr766, and Lys721 as the most common amino acids. In addition to producing the interaction types and numbers from PLIP, it also generates a .pse file to see the 3D conformation of the ligand and its interaction with the protein (.
Figure 15. PLIP analysis of the EGFR-5b complex for each cluster representative. HB: blue solid line, HI: dashed grey line, amino acids: blue sticks, and 5b: orange sticks.
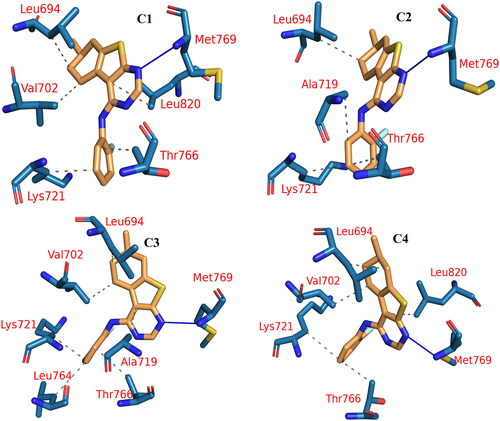
Table 6. The number and types of interactions detected from the PLIP webserver.
ADMET analysis
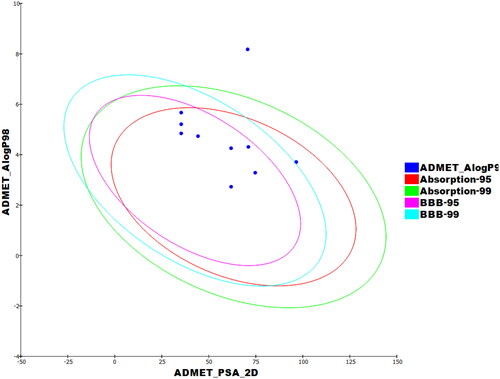
Since the approval of a novel medicine is based on its pharmacokinetic assessment and its biological activity, it is important to explore the pharmacokinetic qualities of a new chemical early on in the drug discovery process to prevent delays in approval or retractionCitation51. Using erlotinib as a reference molecule, the ADMET characteristics of the synthesised compounds were evaluated computationally using Discovery Studio 4.0 ( and ). Although most compounds were predicted to have bad levels of aqueous solubility (Sol. L), most of them expressed good levels of intestinal absorption (Abs. L). Most of the tested compounds were anticipated to have high or very high levels of BBB penetration levels. Regarding the cytochrome P4502D6 inhibitory effects, most compounds were predicted as non-inhibitors. The level of plasma protein binding (PPB. L) model predicts that most compounds were expected to bind plasma protein over 90%.
Table 7. Calculated ADMET descriptors.
Toxicity profiles analysis
For a successful drug development to be minimised the estimation of toxicity is of the utmost importance early on in the processCitation51. In addition, the use of in silico approaches played an indispensable role in the development of drugs to avoid ethical regulations and wasting resources and time on the usual in vitro ad in vivo studiesCitation52. An in silico toxicity prediction, or SAR-predictive toxicity, compares the basic chemical structural properties of molecules being tested with the structures of thousands of compounds with reported safety and toxicityCitation53.
According to the toxicity models built in Discovery Studio software, seven models of toxicity were estimated computationally. These models are; median carcinogenic dose TD50 (MC- TD50) in rats, Maximum Tolerated Dose, in Rats by Feeding (MTDRF), Developmental Toxicity Potential (DTP), Rat Oral LD50 (RLD50), Chronic Lowest-Observed-Adverse-Effect Level (CLOAEL) in addition to the potentialities to be irritant against eye (OI) and skin (SI). As shown in , most compounds showed in silico low toxicity profile against the tested models. Interestingly, compound 5b was predicted to have safer patterns than that of erlotinib in two models (MC-TD50 and MTDRF). Simultaneously, compound 5b was predicted to have almost similar safety patterns to that of erlotinib in four models (CLOAEL, DTP OI, and SI). On the other hand, it was less safe in the model of RLD50.
Table 8. Toxicity properties of the synthesised compounds.
Conclusion
As anti-proliferative lead compounds, a series of thieno[2,3-d] pyrimidine-derived EGFR inhibitors was designed, synthesised, and examined. The most active member, 5b inhibited MCF-7, and A549 cell lines (IC50 = 22.66, and 17.79 µM, respectively). The inhibitory partialities of 5b against two isoforms: EGFRWT and EGFRT790M were assessed to be 37.19, and 204.10 nM, respectively. Interestingly, 5b was much safer (2.5-fold) against the normal cell lines, WI-38, (IC50 =70.13 µM) than erlotinib (IC50 =28.48 µM). Regarding the mechanism of 5b’s anticancer activities, it showed significant potentialities to induce early and late apoptosis in A549 cells based on annexin V staining and arrested the G1 and G2/M phases of A549 cells. To better comprehend and explain the obtained activities on computational levels, molecular docking, MD simulations, MM-GPSA, and PLIP studies against the EGFR protein were performed and confirmed the accurate binding. Therefore, this study presents compound 5b as a potential lead anticancer and EGFR inhibitor.
Experimental
Chemistry
All materials and apparatus used in the synthesis and analyses of the synthesised compounds were clarified in Supplementary Material. Compounds 2 and 3 were synthesised according to the reported proceduresCitation54,Citation55.
4-Chloro-7-methyl-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidine (4)
A mixture of thienopyrimidone 3 (1.1 g, 5 mmol), phosphorus oxychloride (15 ml) and pyridine (1 ml) was heated under reflux for 5 h. The oily residue was poured portion wise into ice cold water (100 ml), and the formed solid was dried and crystallised from ethanol.
Yellow solid (0.9 g, 80%). MP = 100–102 °C; IR (KBr, cm−1): 3059 (CH aromatic), 2954 (CH aliphatic), 1561 (C = N), 1492 (C = C) cm−1; 1HNMR (400 MHz, DMSO-d6, δ ppm) δ 8.79 (s, 1H, C-2H), 3.13–3.12 (m, 2H, CH2), 2.69–2.64 (m, 2H, CH2), 2.21–2.14 (m, 2H, CH2), 1.45–1.42 (m, 1H, CH), 1.06–1.00 (m, 3H, CH3); 13C NMR (101 MHz, DMSO-d6, δ ppm) δ 152.77 (C-2), 152.25 (C-Cl), 144.45, 139.64, 131.19, 128.25 (ArCs), 30.17, 29.21, 28.67, 25.99 (4 aliphatic Cs), 21.41 (CH3).
General procedure for the synthesis of 7-methyl-N-(substituted-phenyl)-5,6,7,8-tetrahydro [1]benzothieno[2,3-d] pyrimidin-4-amine (5a–c)
To a solution of compound 4 (0.24 g, 1 mmol) in acetonitrile for compound 5b or absolute ethanol for compounds 5a and 5c (20 ml), appropriate aniline derivatives (1.1 mmol) were added and the reaction mixture was heated under reflux for 24 h. The formed precipitate was collected, dried and crystallised from ethanol to give the target compounds 5a–c.
N-(4-Chlorophenyl)-7-methyl-5,6,7,8-tetrahydrobenzo[4,5]thieno[2,3-d]pyrimidin-4-amine (5a)
Buff solid (0.3 g, 85%). MP = 170–172 °C; IR (KBr, cm−1): 3450 (NH), 3112 (CH aromatic), 2927 (CH aliphatic), 1662 (C = N), 1563, 1439 (C = C) cm−1; 1H NMR (400 MHz, DMSO-d6, δ ppm) δ 8.40 (s, 1H, C2-H), 8.23 (s, 1H, NH, D2O exchangeable), 7.71 (d, J = 8.4 Hz, 2H, 4-Cl-C6H4-C2,6-H), 7.40 (d, J = 8.4 Hz, 2H, 4-Cl-C6H4-C3,5-H), 3.21–3.18 (m, 2H, CH2), 2.93–2.88 (m, 1H, CH), 2.46–2.39 (m, 1H, CH), 1.94 (d, J = 8 Hz, 2H, CH2), 1.46 (s, 1H, CH), 1.08 (d, J = 8 Hz, 3H, CH3); 13C NMR (101 MHz, DMSO-d6, δ ppm) δ 166.55 (C-2), 155.04 (C-4), 152.42 (C-Cl), 138.77 (C-NH), 133.25, 128.70, 127.33, 126.67, 124.01, 117.27 (ArCs), 33.44, 30.48, 28.82, 25.46 (4 aliphatic Cs), 21.48 (CH3). APCI-MS m/z: Exact mass calcd for C17H16ClN3S [M]+: 329.1. Found: 329.9.
N-(2-Fluorophenyl)-7-methyl-5,6,7,8-tetrahydrobenzo[4,5]thieno[2,3-d]pyrimidin-4-amine (5b)
Reddish brown solid (0.2 g, 70%). MP = 125–127 °C; IR (KBr, cm−1): 3455 (NH), 3090 (CH aromatic), 2951, 2927 (CH aliphatic), 1623 (C = N), 1506, 1450 (C = C) cm−1; 1H NMR (400 MHz, DMSO-d6, δ ppm) δ 8.34 (s, 1H, C2-H), 8.12 (s, 1H, NH, D2O exchangeable), 7.84 (t, J = 8 Hz, 1H, 2-F-C6H4-C4-H), 7.30 (d, J = 8 Hz, 1H, 2-F-C6H4-C3-H), 7.26 (d, J = 4 Hz, 1H, 2-F-C6H4-C6-H), 7.22 (t, J = 8 Hz, 1H, 2-F-C6H4-C5-H), 3.21–3.17 (m, 1H, CH), 3.09–3.07 (m, 1H, CH), 2.93–2.88 (m, 1H, CH), 2.46–2.40 (m, 1H, CH), 1.97–1.94 (m, 2H, CH2), 1.54–1.44 (m, 1H, CH), 1.08 (d, J = 8 Hz, 3H, CH3), 13C NMR (101 MHz, DMSO-d6, δ ppm) δ 166.32 (C-2), 157.46 (C-4), 155.55 (C-F), 155.02 (C-NH), 152.67, 133.14, 127.47, 126.85, 126.59, 124.79, 116.95, 115.87 (ArCs), 33.39, 30.47, 28.80, 25.64 (4 aliphatic Cs), 21.48 (CH3). APCI-MS m/z: Exact mass calcd for C17H16FN3S [M]+: 313.1. Found: 313.8.
N-(4-Methoxyphenyl)-7-methyl-5,6,7,8-tetrahydrobenzo[4,5]thieno[2,3-d]pyrimidin -4-amine (5c)
Violet solid (0.2 g, 62%). MP = 140–142 °C; IR (KBr, cm−1): 3450 (NH), 3101 (CH aromatic), 2951, 2835 (CH aliphatic), 1606 (C = C), 1564, 1443 (C = N) cm−1. 1H NMR (400 MHz, DMSO-d6, δ ppm) δ 8.64 (s, 1H, C2-H), 8.44 (s, 1H, NH, D2O exchangeable), 7.49 (d, J = 8 Hz, 2H, 4-OCH3-C6H4-C3,5-H), 6.99 (d, J = 8 Hz, 2H, 4-OCH3-C6H4-C2,6-H), 3.78 (s, 3H, OCH3), 3.23–3.14 (m, 2H, CH2), 2.96–2.92 (m, 1H, CH), 2.48–2.42 (m, 1H, CH), 1.94 (s, 2H, CH2), 1.51–1.47 (s, 1H, CH), 1.09 (d, J = 8 Hz, 3H, CH3), 13C NMR (101 MHz, DMSO-d6, δ ppm) δ 157.24 (C-2), 155.58 (C-4), 150.17 (C-OCH3), 133.76 (C-NH), 127.26, 126.27, 124.98, 116.74, 115.29, 114.34 (ArCs), 55.81 (OCH3), 33.32, 30.33, 28.74, 25.49 (4 aliphatic Cs), 21.44 (CH3). APCI-MS m/z: Exact mass calcd for C17H17N3OS [M]+: 311.1. Found: 314.1.
N1-(7-Methyl-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4-yl)benzene-1,4-diamine (5d)
4-Chloro-7-methyl-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidine (4) (0.24 g, 1 mmol) was added to a solution of p-phenylenediamine (0.108 g, 1 mmol) in isopropanol (25 ml). The reaction mixture was stirred at 100 °C for 3 h. After cooling, the reaction mixture was poured onto crushed ice with continuous stirring. The formed solid was filtered off, washed with water then dried and crystallised from isopropanol. Gray solid (0.2 g, 65%). MP = 170–172 °C; IR (KBr, cm−1): 3421, 3351 (NH2, NH), 3119 (CH aromatic). 2947, 2922 (CH aliphatic), 1624 (C = N), 1559, 1481 (C = C) cm−1; 1H NMR (400 MHz, DMSO-d6, δ ppm) δ 8.27 (s, 1H, C2-H), 7.85 (s,1H, NH, D2O exchangeable), 7.26 (d, J = 7.9 Hz, 2H, 4-NH2-C6H4-C3,5-H), 6.65 (d, J = 7.8 Hz, 2H, 4-NH2-C6H4-C2,6-H), 5.79 (s, 2H, NH2, D2O exchangeable), 3.18–3.09 (m, 2H, CH2), 2.93–2.86 (s, 1H, CH), 2.45–2.41 (m, 1H, CH), 1.95 (s, 2H, CH2), 1.47 (s, 1H, CH), 1.10 (d, J = 10 Hz, 3H, CH3), 13C NMR (101 MHz, DMSO-d6, δ ppm) δ 165.72 (C-2), 156.16 (C-4), 152.80, 145.82 (C-NH), 131.92 (C-NH2), 128.18, 126.77, 125.53, 116.28, 114.26 (ArCs), 33.41, 30.58, 28.82, 25.70 (4 aliphatic Cs), 21.51 (CH3). APCI-MS m/z: Exact mass calcd for C17H18N4S [M]+: 310.1. Found: 311.2.
General procedure for the synthesis of N-substituted-((7-methyl-5,6,7,8-tetrahydro [1]benzothieno[2,3-d]pyrimidin-4-yl) amino)benzenesulphonamide (5e,f)
Chloropyrimidine 4 (0.24 g, 1 mmol) and the appropriate substituted benzenesulphonamide (1 mmol) in isopropanol (25 ml) was heated under reflux for 10 h. The reaction mixture was allowed to cool, the solid formed was filtered, dried and crystallised from absolute ethanol to give compounds 5e,f.
2-((7-Methyl-5,6,7,8-tetrahydrobenzo[4,5]thieno[2,3-d]pyrimidin-4-yl)amino)benzene sulphonamide (5e)
Buff solid (0.3 g, 83%). MP = 200–202 °C; IR (KBr, cm−1): 3482, 3391, 3251 (NH2, NH), 3052 (CH aromatic), 2925 (CH aliphatic), 1617 (C = N), 1532, 1414 (C = C) cm−1; 1H NMR (400 MHz, DMSO-d6, δ ppm) δ 8.81 (s, 1H, C2-H), 7.54 (d, J = 8 Hz, 1H, 2-H2NO2S-C6H4-C-3-H), 7.23 (t, J = 8 Hz 1H, 2-H2NO2S-C6H4-C-4-H), 7.11 (s, 1H, NH, D2O exchangeable), 6.79 (d, J = 8 Hz, 1H, 2-H2NO2S-C6H4-C-6-H), 6.60 (t, J = 8 Hz, 1H, 2-H2NO2S-C6H4-C-5-H), 5.60 (s, 2H, NH2, D2O exchangeable), 3.21–3.17 (m, 2H, CH2), 3.02–2.92 (m. 2H, CH2), 1.96 (d, J = 8 Hz, 2H, CH2), 1.51–1.45 (m, 1H, CH), 1.08 (d, J = 8 Hz, 3H, CH3). 13C NMR (101 MHz, DMSO-d6, δ ppm) δ 168.66 (C-2), 157.07 (C-4), 152.82, 152.29, 146.02 (C-NH), 139.70, 133.32 (C-SO2-NH2), 128.34, 126.81, 124.68, 117.15, 115.42, (ArCs), 33.68, 30.18, 28.68, 25.99 (4 aliphatic Cs), 21.41 (CH3).
4-((7-Methyl-5,6,7,8-tetrahydrobenzo[4,5]thieno[2,3-d]pyrimidin-4-yl)amino)benzene sulphonamide (5f)
Buff solid (0.3 g, 79%). MP = 210–212 °C; IR (KBr, cm−1): 3436, 3325, 3200 (NH2, NH), 3040 (CH aromatic), 2956 (CH aliphatic), 1604 (C = N), 1562, 1439 (C = C) cm−1; 1H NMR (400 MHz, DMSO-d6, δ ppm) δ 8.51 (s, 1H, C2-H), 8.48 (s, 1H, NH, D2O exchangeable), 7.85 (d, J = 8.8 Hz, 2H, 4-H2NO2S-C6H4-C-2,6-H), 7.79 (d, J = 8.8 Hz, 2H, 4-H2NO2S-C6H4-C-3,5-H), 7.28 (s, 2H, NH2, D2O exchangeable), 3.25–3.18 (m, 2H, CH2), 2.97–2.92 (m, 1H, CH), 2.47–2.43 (m, 1H, CH), 1.98–1.95 (m, 2H, CH2), 1.54–1.44 (m, 1H, CH), 1.10 (d, J = 8 Hz, 3H, CH3); 13C NMR (101 MHz, DMSO-d6, δ ppm) δ 166.93 (C-2), 161.61 (C-4), 154.74, 152.37, 143.01 (C-NH), 138.34, 133.84 (C-SO2-NH2), 126.69, 121.24, 117.87 (ArCs), 33.47, 30.45, 28.83, 25.36 (4 aliphatic Cs), 21.47 (CH3).
N-((7-Methyl-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4-yl)methyl) aniline (6)
A solution of chloropyrimidine 4 (0.24 g, 1 mmol) in absolute ethanol (25 ml) was treated with benzylamine (0.12 g, 1.1 mmol) and heated under reflux for 24 h. The solid obtained after cooling was dried and crystallised from isopropanol.
Buff solid (0.2 g, 64%) MP = 140–142 °C; IR (KBr, cm−1): 3403 (NH), 3052 (CH aromatic), 2933, 2852 (CH aliphatic), 1610 (C = N), 1510, 1445 (C = C) cm−1; 1H NMR (400 MHz, DMSO-d6, δ ppm) δ 8.81 (s, 1H, C2-H), 8.25 (s, 5H, ArH), 5.87 (s, 1H, NH, D2O exchangeable, 4.09 (s, 2H, CH2-NH), 3.01–2.97 (m, 1H, CH), 2.85–2.81 (m, 1H, CH), 2.39–2.33 (m, 1H, CH), 1.72 (d, J = 8 Hz, 1H, CH), 1.61 (d, J = 8 Hz, 2H, CH2), 1.23–1.17 (m, 1H, CH), 1.05 (d, J = 4 Hz, 3H, CH3). 13C NMR (101 MHz, DMSO-d6, δ ppm) δ 165.13 (C-2), 156.68 (C-4), 153.20, 152.27, 139.65 (C-NH), 131.29, 128.31, 126.61, 115.69 (ArCs), 49.46 (CH2-NH), 33.26, 30.48, 28.73, 25.19, (4 aliphatic Cs), 21.45 (CH3). APCI-MS m/z: Exact mass calcd for C18H19N3S [M]+: 309.1. Found: 309.1.
2–(7-Methyl-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4-yl)hydrazine carbothioamide (7a)
Chloropyrimidine 4 (0.24 g, 1 mmol) and thiosemicarbazide (0.1 g, 1 mmol) was dissolved in absolute ethanol (25 ml) and heated under reflux for 8 h. The precipitated solid was filtered, dried and crystallised from ethanol.
Orange solid (0.3 g, 82%). MP = 213–215 °C; IR (KBr, cm−1): 3494, 3428, 3301 (NH2, two NHs), 3077 (CH aromatic), 2944 (CH aliphatic), 1647 (C = N), 1536, 1460 (C = C) cm−1; 1H NMR (400 MHz, DMSO-d6, δ ppm) δ 9.17 (s, 1H, C2-H), 8.44 (s, 1H, NH, D2O exchangeable), 8.10 (s, 1H, NH, D2O exchangeable), 6.49 (s, 2H, NH2, D2O exchangeable), 3.17–3.13 (m, 1H, CH), 2.97–2.88 (m, 2H, CH2), 2.46–2.42 (m, 1H, CH), 1.98–1.89 (m, 2H, CH2), 1.49–1.48 (m, 1H, CH), 1.09 (d, J = 6.6 Hz, 3H, CH3); 13C NMR (101 MHz, DMSO-d6, δ ppm) δ 178.51 (C = S), 167.16 (C-2), 153.55 (C-4), 148.71 (C-NH), 136.42, 135.89, 128.08 (ArCs), 33.20, 30.39, 29.31, 25.19 (4 aliphatic Cs), 21.56 (CH3). APCI-MS m/z: Exact mass calcd for C12H15N5S2 [M]+: 293.08.
4-Hydrazinyl-7-methyl-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidine (7b)
Hydrazine hydrate (0.1 g, 2 mmol, 99%) solution in absolute ethanol (25 ml) was added to chloropyrimidine 4 (0.24 g, 1 mmol) and heated under reflux for 5 h. After cooling, the mixture was poured on ice-cooled water, filtered, dried and crystallised from ethanol to give the hydrazinyl derivative 7b.
Buff solid (0.2 g, 58%). MP = 180–182 °C; IR (KBr, cm−1): 3421, 3346 (NH2, NH), 3082 (CH aromatic), 2922, 2861 (CH aliphatic), 1636 (C = N), 1560, 1473 (C = C) cm−1; 1H NMR (400 MHz, DMSO-d6, δ ppm) δ 8.33 (s, 1H, C2-H), 7.87 (s, 1H, NH, D2O exchangeable), 4.65 (s, 2H, NH2, D2O exchangeable), 3.03–2.81 (m, 2H, CH2), 2.40–2.34 (m, 2H, CH2), 1.90–1.87 (m, 2H, CH2), 1.43–1.39 (m, 1H, CH), 1.05 (d, J = 8 Hz, 3H, CH3); 13C NMR (101 MHz, DMSO-d6, δ ppm) δ 164.49 (C-4), 158.68 (C-2), 152.93, 131.56, 126.95, 115.17 (ArCs), 33.30, 30.60, 28.85, 25.61 (4 aliphatic Cs), 21.51 (CH3).
NCitation1,N-4-Bis(7-methyl-5,6,7,8-tetrahydrobenzo[4,5]thieno[2,3-d]pyrimidin-4-yl) benzene-1,4-diamine (8)
A solution of chloropyrimidine 4 (0.24 g, 1 mmol) and p-phenylenediamine (0.12 g, 1.1 mmol) in isopropanol (25 ml) was heated under reflux for 6 h with stirring. The mixture was filtered on hot, washed with ethanol and dried to give the dimer 8.
Buff solid (0.3 g, 55%). MP = 220–222 °C; IR (KBr, cm−1): 3439, 3400 (two NHs), 3020 (CH aromatic), 2949, 2918 (CH aliphatic), 1585 (C = N), 1443 (C = C) cm−1; 1H NMR (400 MHz, DMSO-d6, δ ppm) δ 8.36 (s, 2H, two C2-H), 8.11 (s, 2H, two NHs, D2O exchangeable), 7.61 (s, 4H, ArH), 3.25–3.15 (m, 4H, two CH2), 2.94–2.90 (m, 2H, two CH), 2.51–2.41 (m, 2H, two CH), 1.98–1.95 (m, 4H, two CH2), 1.51–1.49 (m, 2H, two CH), 1.09 (d, J = 4 Hz, 6H, two CH3); 13C NMR (101 MHz, DMSO-d6, δ ppm) δ 174.44 (C-4), 165.85 (C-2), 157.76, 152.75, 132.17, 126.77, 125.20, 123.19, 116.42, (ArCs), 33.41, 30.53, 28.81, 25 0.95 (4 aliphatic Cs), 21.48 (CH3); APCI-MS m/z: Exact mass calcd for C28H28N6S2 [M]+: 512.1. Found: 511.1.
Biological examinations
In vitro anti-proliferative activity
The anti-proliferative activities of the synthesised compounds against A549 and MCF-7cell lines were carried out using an MTT procedure as shown in Supplementary MaterialCitation42,Citation56,Citation57.
In vitro EGFR inhibition
The inhibitory activities of the synthesised compounds against EGFRWT and EGFRT790M proteins were carried out using EGFR Kinase Assay Kit (BPS Bioscience, USA) as shown in Supplementary MaterialCitation47.
Safety assay
The cytotoxic effect of compound 5b against a normal cell line (W138) was assessed using an MTT assay.
Cell cycle analysis
The cell cycle analysis was carried out using a flow cytometer as shown in Supplementary MaterialCitation58,Citation59.
Apoptosis analysis
The apoptotic effect of compound 5b was tested using a flow cytometer as shown in Supplementary MaterialCitation60,Citation61.
BAX and Bcl-2 determination
Supplementary Material provides a thorough justification. The effect of compound 5b on BAX and Bcl-2 level in A549 cellsCitation62–64.
In silico studies
Docking studies
Docking studies against EGFRWT and EGFRT790M were performed by MOE2019 software as shown in Supplementary MaterialCitation35.
MD simulations
MD simulation studies were directed by CHARMM-GUI web server and GROMACS 2021 as an MD engine as shown in Supplementary MaterialCitation65,Citation66.
MM-GBSA and PLIP
MM-GBSA and PLIP analyses were performed by the Gmx_MMPBSA package as shown in Supplementary MaterialCitation67.
ADMET studies
ADMET profile was examined by Discovery Studio 4.0 as shown in Supplementary MaterialCitation68.
Toxicity studies
The toxicity profile was tested by Discovery Studio 4.0 as shown in Supplementary MaterialCitation69.
Supplemental Material
Download PDF (11.6 MB)Acknowledgement
The authors extend their appreciation to the Research Center at AlMaarefa University for funding this work.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249.
- WHO. Cancer, Overview. [accessed 2022 Oct 1]. https://www.who.int/health-topics/cancer#tab=tab_1.
- Badran MM, Abouzid KM, Hussein MHM. Synthesis of certain substituted quinoxalines as antimicrobial agents (part II). Arch Pharm Res. 2003;26(2):107–113.
- WHO. Cancer, Key facts. [accessed 2023 Mar 1]. https://www.who.int/news-room/fact-sheets/detail/cancer.
- Chabner BA, Roberts TG. Chemotherapy and the war on cancer. Nat Rev Cancer. 2005;5(1):65–72.
- Krauss G, Schönbrunner N, Cooper J. Biochemistry of signal transduction and regulation. Weinheim (Germany): Wiley Online Library; 2003. Vol. 3.
- Nguyen K-SH, Kobayashi S, Costa DB. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non–small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin Lung Cancer. 2009;10(4):281–289.
- Grant S. Therapeutic protein kinase inhibitors. Cell Mol Life Sci. 2009;66(7):1163–1177.
- Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, De Feo G, Caponigro F, Salomon DS, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366(1):2–16.
- Huang S-M, Harari PM. Epidermal growth factor receptor inhibition in cancer therapy: biology, rationale and preliminary clinical results. Invest New Drugs. 1999;17(3):259–269.
- Nicholson RI, Gee JMW, Harper ME. egfR and cancer prognosis. Eur J Cancer. 2001;37:9–15.
- Zhang H-Q, Gong F-H, Ye J-Q, Zhang C, Yue X-H, Li C-G, Xu Y-G, Sun L-P. Design and discovery of 4-anilinoquinazoline-urea derivatives as dual TK inhibitors of EGFR and VEGFR-2. Eur J Med Chem. 2017;125:245–254.
- Song Z, Huang S, Yu H, Jiang Y, Wang C, Meng Q, Shu X, Sun H, Liu K, Li Y, et al. Synthesis and biological evaluation of morpholine-substituted diphenylpyrimidine derivatives (Mor-DPPYs) as potent EGFR T790M inhibitors with improved activity toward the gefitinib-resistant non-small cell lung cancers (NSCLC). Eur J Med Chem. 2017;133:329–339.
- Zhang Y, Chen L, Xu H, Li X, Zhao L, Wang W, Li B, Zhang X. 6, 7-Dimorpholinoalkoxy quinazoline derivatives as potent EGFR inhibitors with enhanced antiproliferative activities against tumor cells. Eur J Med Chem. 2018;147:77–89.
- Bonomi P. Erlotinib: a new therapeutic approach for non-small cell lung cancer. Expert Opin Investig Drugs. 2003;12(8):1395–1401.
- Ou S-HI. Second-generation irreversible epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs): a better mousetrap? A review of the clinical evidence. Crit Rev Oncol Hematol. 2012;83(3):407–421.
- Walter AO, Sjin RTT, Haringsma HJ, Ohashi K, Sun J, Lee K, Dubrovskiy A, Labenski M, Zhu Z, Wang Z, et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov. 2013;3(12):1404–1415.
- Sequist LV, Besse B, Lynch TJ, Miller VA, Wong KK, Gitlitz B, Eaton K, Zacharchuk C, Freyman A, Powell C, et al. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non–small-cell lung cancer. J Clin Oncol. 2010;28(18):3076–3083.
- Kim Y, Ko J, Cui ZYun, Abolhoda A, Ahn JS, Ou S-H, Ahn M-J, Park K. The EGFR T790M mutation in acquired resistance to an irreversible second-generation EGFR inhibitor. Mol Cancer Ther. 2012;11(3):784–791.
- Zhao Z, Wu H, Wang L, Liu Y, Knapp S, Liu Q, Gray NS. Exploration of type II binding mode: a privileged approach for kinase inhibitor focused drug discovery? ACS Chem Biol. 2014;9(6):1230–1241.
- Furet P, Caravatti G, Lydon N, Priestle JP, Sowadski JM, Trinks U, Traxler P. Modelling study of protein kinase inhibitors: binding mode of staurosporine and origin of the selectivity of CGP 52411. J Comput Aided Mol Des. 1995;9(6):465–472.
- Gandin V, Ferrarese A, Dalla Via M, Marzano C, Chilin A, Marzaro G. Targeting kinases with anilinopyrimidines: discovery of N-phenyl-N’-[4-(pyrimidin-4-ylamino) phenyl] urea derivatives as selective inhibitors of class III receptor tyrosine kinase subfamily. Sci Rep. 2015;5:16750.
- Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol. 2006;2(7):358–364.
- Elrazaz EZ, Serya RAT, Ismail NSM, Abou El Ella DA, Abouzid KAM. Thieno [2, 3-d] pyrimidine based derivatives as kinase inhibitors and anticancer agents. Future J Pharm Sci. 2015;1(2):33–41.
- Yang C-R, Peng B, Cao S-L, Ren T-T, Jiang W, Wang F-C, Li Y-S, Wang G, Li Z, Xu S, et al. Synthesis, cytotoxic evaluation and target identification of thieno [2, 3-d] pyrimidine derivatives with a dithiocarbamate side chain at C2 position. Eur J Med Chem. 2018;154:324–340.
- Rashad A, Ali M. Synthesis and antiviral screening of some thieno [2, 3-d] pyrimidine nucleosides. Nucleosides Nucleotides Nucleic Acids. 2006;25(1):17–28.
- Kotaiah Y, Harikrishna N, Nagaraju K, Venkata Rao C. Synthesis and antioxidant activity of 1, 3, 4-oxadiazole tagged thieno [2, 3-d] pyrimidine derivatives. Eur J Med Chem. 2012;58:340–345.
- Vega S, Alonso J, Diaz JA, Junquera F, Perez C, Darias V, Bravo L, Abdallah S. Thiophene isosteres: synthesis and biological evaluation of 3-substituted derivatives of 4-phenyl-2-thioxo-benzo [4, 5] thieno [2, 3-d] pyrimidine. Eur J Med Chem . 1991;26(3):323–329.
- El-Gazzar A-R, Hussein H, Hafez H. Synthesis and biological evaluation of thieno [2, 3-d] pyrimidine derivatives for anti-inflammatory, analgesic and ulcerogenic activity. Acta Pharm. 2007;57(4):395–411.
- Vlasov SV, Osolodchenko TP, Kovalenko SM, Chernykh VP. Synthesis and the antimicrobial activity of 1-alkyl-5-methyl-3-phenyl-6-(5-phenyl-1, 3, 4-oxadiazol-2-yl) thieno [2, 3-d] pyrimidine-2, 4 (1H, 3H)-diones. J Org Pharm Chem. 2015;13(1(49)):20–24.
- Ali EMH, Abdel-Maksoud MS, Oh C-H. [2, 3-d] pyrimidine as a promising scaffold in medicinal chemistry: Recent advances. Bioorg Med Chem. 2019;27(7):1159–1194.
- Bugge S, Kaspersen SJ, Larsen S, Nonstad U, Bjørkøy G, Sundby E, Hoff BH. Structure–activity study leading to identification of a highly active thienopyrimidine based EGFR inhibitor. Eur J Med Chem. 2014;75:354–374.
- Gaber AA, Bayoumi AH, El-Morsy AM, Sherbiny FF, Mehany ABM, Eissa IH. Design, synthesis and anticancer evaluation of 1H-pyrazolo [3, 4-d] pyrimidine derivatives as potent EGFRWT and EGFRT790M inhibitors and apoptosis inducers. Bioorg Chem. 2018;80:375–395.
- Elmetwally SA, Saied KF, Eissa IH, Elkaeed EB. Design, synthesis and anticancer evaluation of thieno [2, 3-d] pyrimidine derivatives as dual EGFR/HER2 inhibitors and apoptosis inducers. Bioorg Chem. 2019;88:102944.
- Nasser AA, Eissa IH, Oun MR, El-Zahabi MA, Taghour MS, Belal A, Saleh AM, Mehany ABM, Luesch H, Mostafa AE, et al. Discovery of new pyrimidine-5-carbonitrile derivatives as anticancer agents targeting EGFR WT and EGFR T790M. Org Biomol Chem. 2020;18(38):7608–7634.
- Zhao Z, Xie L, Bourne PE. Structural insights into characterizing binding sites in epidermal growth factor receptor kinase mutants. J Chem Inf Model. 2019;59(1):453–462.
- Kandeel MM, Mounir AA, Refaat HM, Kassab AE. Synthesis of thieno [2, 3-d] pyrimidines, thieno [2, 3-d] triazinones and thieno [2, 3-e] diazepinones of anticipated anti-cancer activity. J Chem Res. 2012;36(2):105–110.
- Tormyshev V, Trukhin D, Rogozhnikova O, Mikhalina T, Troitskaya T, Flinn A. Aryl alkyl ketones in a one-pot Gewald synthesis of 2-aminothiophenes. Synlett. 2006;2006(16):2559–2564.
- El‐Adl K, Ibrahim M‐K, Khedr F, Abulkhair HS, Eissa IH. N‐Substituted‐4‐phenylphthalazin‐1‐amine‐derived VEGFR‐2 inhibitors: design, synthesis, molecular docking, and anticancer evaluation studies. Arch Pharm. 2021;354(3):2000219.
- Abdelhaleem EF, Abdelhameid MK, Kassab AE, Kandeel MM. Design and synthesis of thienopyrimidine urea derivatives with potential cytotoxic and pro-apoptotic activity against breast cancer cell line MCF-7. Eur J Med Chem. 2018;143:1807–1825.
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1-2):55–63.
- Denizot F, Lang R. Rapid colorimetric assay for cell growth and survival: modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J Immunol Methods. 1986;89(2):271–277.
- Thabrew MI, Hughes RD, Mcfarlane IG. Screening of hepatoprotective plant components using a HepG2 cell cytotoxicity assay. J Pharm Pharmacol. 1997;49(11):1132–1135.
- Li P, Zhang Q, Torossian A, Li Z-b, Xu W-c, Lu B, Fu S. Simultaneous inhibition of EGFR and PI3K enhances radiosensitivity in human breast cancer. Int J Radiat Oncol Biol Phys. 2012;83(3):e391–e397.
- Huether A, Höpfner M, Baradari V, Schuppan D, Scherübl H. EGFR blockade by cetuximab alone or as combination therapy for growth control of hepatocellular cancer. Biochem Pharmacol. 2005;70(11):1568–1578.
- Van Schaeybroeck S, Kyula J, Kelly DM, Karaiskou-McCaul A, Stokesberry SA, Van Cutsem E, Longley DB, Johnston PG. Chemotherapy-induced epidermal growth factor receptor activation determines response to combined gefitinib/chemotherapy treatment in non–small cell lung cancer cells. Mol Cancer Ther. 2006;5(5):1154–1165.
- Jia Y, Quinn CM, Gagnon AI, Talanian R. Homogeneous time-resolved fluorescence and its applications for kinase assays in drug discovery. Anal Biochem. 2006;356(2):273–281.
- Pietenpol J, Stewart Z. Cell cycle checkpoint signaling:: cell cycle arrest versus apoptosis. Toxicology. 2002;181-182:475–481.
- Park JH, Liu Y, Lemmon MA, Radhakrishnan R. Erlotinib binds both inactive and active conformations of the EGFR tyrosine kinase domain. Biochem J. 2012;448(3):417–423.
- Sogabe S, Kawakita Y, Igaki S, Iwata H, Miki H, Cary DR, Takagi T, Takagi S, Ohta Y, Ishikawa T, et al. Structure-based approach for the discovery of pyrrolo [3, 2-d] pyrimidine-based EGFR T790M/L858R mutant inhibitors. ACS Med Chem Lett. 2013;4(2):201–205.
- Ferreira LL, Andricopulo A. ADMET modeling approaches in drug discovery. Drug Discov Today. 2019;24(5):1157–1165.
- Dearden JC. In silico prediction of drug toxicity. J Comput Aided Mol Des. 2003;17(2-4):119–127.
- Kruhlak NL, Benz RD, Zhou H, Colatsky TJ. (Q) SAR modeling and safety assessment in regulatory review. Clin Pharmacol Ther. 2012;91(3):529–534.
- Golub AG, Bdzhola VG, Briukhovetska NV, Balanda AO, Kukharenko OP, Kotey IM, Ostrynska OV, Yarmoluk SM. Synthesis and biological evaluation of substituted (thieno[2,3-d]pyrimidin-4-ylthio)carboxylic acids as inhibitors of human protein kinase CK2. Eur J Med Chem. 2011;46(3):870–876.
- Abbas SE, Abdel Gawad NM, George RF, Akar YA. Synthesis, antitumor and antibacterial activities of some novel tetrahydrobenzo[4,5]thieno[2,3-d]pyrimidine derivatives. Eur J Med Chem. 2013;65:195–204.
- Al-Rashood ST, Hamed AR, Hassan GS, Alkahtani HM, Almehizia AA, Alharbi A, Al-Sanea MM, Eldehna WM. Antitumor properties of certain spirooxindoles towards hepatocellular carcinoma endowed with antioxidant activity. J Enzyme Inhib Med Chem. 2020;35(1):831–839.
- Elkady H, Elwan A, El-Mahdy HA, Doghish AS, Ismail A, Taghour MS, Elkaeed EB, Eissa IH, Dahab MA, Mahdy HA, et al. New benzoxazole derivatives as potential VEGFR-2 inhibitors and apoptosis inducers: design, synthesis, anti-proliferative evaluation, flowcytometric analysis, and in silico studies. J Enzyme Inhib Med Chem. 2022;37(1):403–416.
- Wang J, Lenardo MJ. Roles of caspases in apoptosis, development, and cytokine maturation revealed by homozygous gene deficiencies. J Cell Sci. 2000;113(5):753–757.
- Eldehna WM, Hassan GS, Al-Rashood ST, Al-Warhi T, Altyar AE, Alkahtani HM, Almehizia AA, Abdel-Aziz HA. Synthesis and in vitro anticancer activity of certain novel 1-(2-methyl-6-arylpyridin-3-yl)-3-phenylureas as apoptosis-inducing agents. J Enzyme Inhib Med Chem. 2019;34(1):322–332.
- Lo KK-W, Lee TK-M, Lau JS-Y, Poon W-L, Cheng S-H. Luminescent biological probes derived from ruthenium (II) estradiol polypyridine complexes. Inorg Chem. 2008;47(1):200–208.
- Sabt A, Abdelhafez OM, El-Haggar RS, Madkour HMF, Eldehna WM, El-Khrisy EE-DAM, Abdel-Rahman MA, Rashed LA. Novel coumarin-6-sulfonamides as apoptotic anti-proliferative agents: synthesis, in vitro biological evaluation, and QSAR studies. J Enzyme Inhib Med Chem. 2018;33(1):1095–1107.
- Balah A, Ezzat O, Akool E-S. Vitamin E inhibits cyclosporin A-induced CTGF and TIMP-1 expression by repressing ROS-mediated activation of TGF-β/Smad signaling pathway in rat liver. Int Immunopharmacol. 2018;65:493–502.
- Aborehab NM, Elnagar MR, Waly NE. Gallic acid potentiates the apoptotic effect of paclitaxel and carboplatin via overexpression of Bax and P53 on the MCF‐7 human breast cancer cell line. J Biochem Mol Toxicol. 2021;35(2):e22638.
- Elnagar MR, Walls AB, Helal GK, Hamada FM, Thomsen MS, Jensen AA. Functional characterization of α7 nicotinic acetylcholine and NMDA receptor signaling in SH-SY5Y neuroblastoma cells in an ERK phosphorylation assay. Eur J Pharmacol. 2018;826:106–113.
- Elkaeed EB, Eissa IH, Elkady H, Abdelalim A, Alqaisi AM, Alsfouk AA, Elwan A, Metwaly AM. A multistage in silico study of natural potential inhibitors targeting SARS-CoV-2 main protease. Int J Mol Sci. 2022;23(15):8407.
- Elkaeed EB, Youssef FS, Eissa IH, Elkady H, Alsfouk AA, Ashour ML, El Hassab MA, Abou-Seri SM, Metwaly AM. Multi-step in silico discovery of natural drugs against COVID-19 targeting main protease. Int J Mol Sci. 2022;23(13):6912.
- Elkaeed EB, Yousef RG, Elkady H, Gobaara IMM, Alsfouk BA, Husein DZ, Ibrahim IM, Metwaly AM, Eissa IH. Design, synthesis, docking, DFT, MD simulation studies of a new nicotinamide-based derivative: in vitro anticancer and VEGFR-2 inhibitory effects. Molecules. 2022;27(14):4606.
- Taghour MS, Elkady H, Eldehna WM, El-Deeb NM, Kenawy AM, Elkaeed EB, Alsfouk AA, Alesawy MS, Metwaly AM, Eissa IH, et al. Design and synthesis of thiazolidine-2, 4-diones hybrids with 1, 2-dihydroquinolones and 2-oxindoles as potential VEGFR-2 inhibitors: in-vitro anticancer evaluation and in-silico studies. J Enzyme Inhib Med Chem. 2022;37(1):1903–1917.
- Yousef RG, Sakr HM, Eissa IH, Mehany ABM, Metwaly AM, Elhendawy MA, Radwan MM, ElSohly MA, Abulkhair HS, El-Adl K, et al. New quinoxaline-2 (1 H)-ones as potential VEGFR-2 inhibitors: Design, synthesis, molecular docking, ADMET profile and anti-proliferative evaluations. New J Chem. 2021;45(36):16949–16964.