?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Non-structural protein 5 (Nsp5) is a cysteine protease that plays a key role in SARS-CoV-2 replication, suppressing host protein synthesis and promoting immune evasion. The investigation of natural products as a potential strategy for Nsp5 inhibition is gaining attention as a means of developing antiviral agents. In this work, we have investigated the physicochemical properties and structure-activity relationships of ellagic acid and its gut metabolites, urolithins A–D, as ligands of Nsp5. Results allow us to identify urolithin D as promising ligand of Nsp5, with a dissociation constant in the nanomolar range of potency. Although urolithin D is able to bind to the catalytic cleft of Nsp5, the appraisal of its viral replication inhibition against SARS-CoV-2 in Vero E6 assay highlights a lack of activity. While these results are discussed in the framework of the available literature reporting conflicting data on polyphenol antiviral activity, they provide new clues for natural products as potential viral protease inhibitors.
Introduction
Coronaviruses are members of the coronaviridae family which includes enveloped, positive single-stranded RNA viruses that infect a wide range of hosts, such as avian, swine, and humans. Until the 21st century, most members of the family caused only mild respiratory effects on humans, but the appearance of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) in 2003 and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in 2012, has increased interest towards these pathogens. In 2019, a novel coronavirus, namely the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), spread causing a world pandemic with a significant impact on global healthcare and economyCitation1,Citation2. These events have posed a medical need for novel therapeutic antiviral agents, fostering the development of drug discovery strategies to identify novel magic bulletsCitation3 targeting the essential components of the viral life cycle.
SARS-CoV-2 single-stranded RNA (ssRNA) genome is translated by host ribosomes into two large polyproteins: pp1a and pp1ab. These polyproteins are cleaved by two cysteine proteases, namely papain-like protease (PLpro or Nsp3) and chymotrypsin-like protease (3CLpro or Nsp5), producing mature structural and non-structural proteins, such as the RNA-dependent RNA polymerase, helicase, and exonuclease that are fundamental components for the viral replicationCitation4. Since there are eleven Nsp5 cleavage sites in the polyproteins, this enzyme is considered the viral main proteaseCitation5, and its substrate specificity is characterised by the recognition and cleavage of an aminoacidic motif substrate (LQ/SAG motif) that is not recognised by human proteasesCitation6. SARS-CoV-2 Nsp5 shares 96.1 and 50.3% sequence identity with the proteases of SARS-CoV and MERS-CoV, respectivelyCitation7.
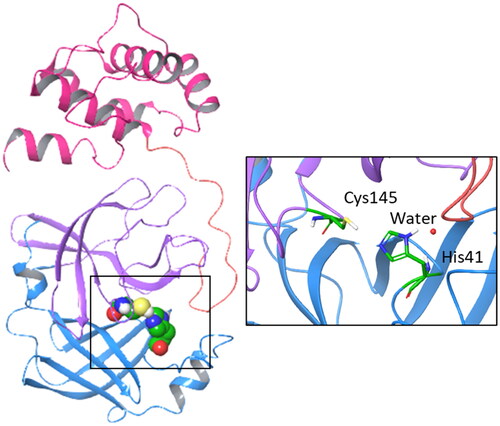
From a structural point of view, Nsp5 is a functional homodimer and each monomer (33.8 kDa) is composed of three distinct domains (). Domains I (8–101 residues) and II (102–184 residues) are formed by six-stranded antiparallel β-barrels, whereas domain III (201–303 residues) is composed of five α-helices which are linked to domain II by a long loop (185–200 residues)Citation8. The active site is located between domains I and II, consisting of a catalytic dyad (Cys145 and His41) and a water molecule that forms a hydrogen bond with His41 thereby acting as a third component of the catalytic machineryCitation8,Citation9. Domain III is involved in the dimerisation process which is required for the catalytic activity of the enzymeCitation10 and affected by the presence of ligandsCitation11.
Figure 1. Structure, domains, and catalytic dyad of the viral main protease Nsp5. Domain I (8–101 residues) is shown with blue ribbon; domain II (102–184 residues) is shown with purple ribbon; domain III (201–303 residues) is shown with magenta ribbon; the loop (185–200 residues) connecting domains II and III is shown with pink ribbon. The catalytic machinery is highlighted in the box.
Due to its pivotal role in regulating the viral life cycle and lack of protein homologues in humans, Nsp5 has been thrust into the spotlight as promising druggable target for the development of antiviral SARS-CoV-2 agentsCitation9. Accordingly, both the design of irreversible peptidomimetic inhibitors and nonpeptidic inhibitors has been pursued as key strategies to block the catalytic activity of the viral main protease Nsp5Citation12,Citation13. The first strategy relies on combining the design of substrate analogues with the insertion of reactive groups (warheads) that are able to form a covalent adduct with the catalytic cysteine residue (Cys145) by undergoing a nucleophilic attack. While Michael acceptor groups are commonly used as warheads, other moieties, such as cyano or trifluoromethyl groups have also been exploited to obtain a reversible covalent bond formation with Cys145 for reducing potential toxic effects that can be associated with the likelihood of off-target activities of the irreversible covalent binders. Nirmatrelvir (1, ) is a successful example of a reversible covalent binder that has been recently approved for the treatment of SARS-CoV-2 diseaseCitation14,Citation15. The design of nonpeptidic inhibitors is based on the identification of small molecules that act as irreversible or reversible inhibitors of the main protease.
Figure 2. Chemical structures and mechanistic profile of Nirmatrelvir (1) and selected inhibitors (2–6) of the viral main protease Nsp5.
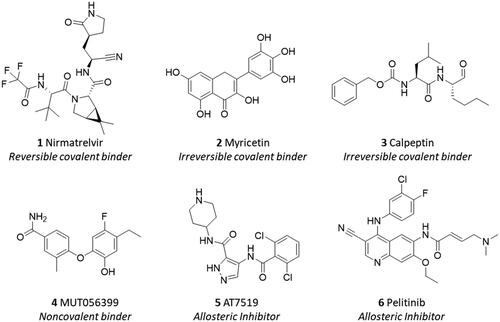
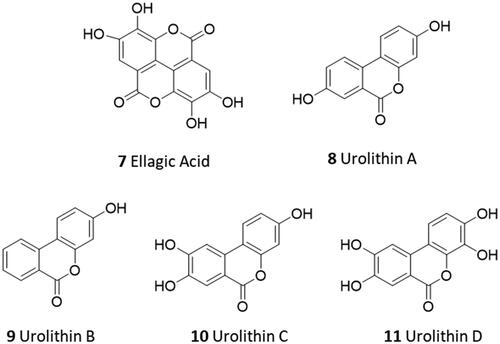
In this regard, screening campaigns of large chemical libraries represent a valuable strategy to identify high-quality hit compounds, including natural products and repurposed drugs with either irreversible, reversible and allosteric inhibitor profiles (e.g. myricetin 2, calpeptin 3, MUT056399 4, AT7519 5, pelitinib 6) that may feed further optimisation efforts on the way to clinical candidatesCitation16,Citation17. In this framework, over the SARS-CoV-2 pandemic years, natural products have attracted considerable attention in the scientific community as a source of promising hit compounds for the development of anti-viral SARS-CoV-2 agents. Several studies have indeed disclosed flavonoids, chalcones, isatin, and terpenoid derivatives as Nsp5 inhibitorsCitation18–26. Part of these studies has also reported dietary hydrolysable tannins as promising inhibitors of the viral main protease Nsp5, including ellagic acid (7) and some of its gut microbiota metabolitesCitation27–29. In some of these works, molecular docking protocols have been applied without experimental validation to support the relative findingsCitation30,Citation31. In some others, discrepancies have been noticed among biochemical findings, biological outcomes, and computational results about natural products as ligands and/or inhibitors of Nps5. For instance, in a recent study, researchers have reported low and high micromolar dissociation constants of ellagic acid (7, ) and its metabolite urolithin B (9) to Nsp5 from surface plasmon resonance (SPR) experiments, whereas an undetectable binding activity was observed for the other metabolite urolithin A (8)Citation28.
These findings agreed only in part with data from computational studies, showing similar favourable free binding energy for such compounds to the catalytic site, and with biological results indicating mild or poor Nsp5 inhibition activity.
In the present work, we deemed of interest to devote further research efforts to determine the structure-activity relationship (SAR) and inhibition activity of ellagic acid (7) and urolithin metabolites (urolithin A, B, C, and D; 8–11) against SARS-CoV-2 main protease Nsp5, using physicochemical characterisations, microscale thermophoresis (MST) assays, computational studies, and cellular appraisals in African green monkey kidney cells (Vero-6 cell line). While MST represents a complementary technique to complement previous SPR experiments providing consensus data, the use of Vero-6 cell line represents a disease-relevant assay for the appraisal of the inhibition activity of these dietary polyphenol derivatives against the viral life cycle of SARS-CoV-2.
Results and discussion
Ellagic acid (7) is a natural compound that is contained in various plants and vegetable species in free form or embedded in polymeric forms termed ellagitanninsCitation32,Citation33. Specifically, ellagitannins release ellagic acid upon hydrolysis, which is further metabolised to urolithins by gut microbiota. Both ellagic acid and urolithins contain the benzo[c]coumarin nucleus as a core scaffold. Although ellagic acid is mainly known for its antioxidant functions, other biological functions have been reported, such as anti-inflammatory, anticancer, and neuroprotective functionsCitation34. It is worth noting that ellagic acid has been indicated as the main phytochemical compound accounting for the antiviral and antibacterial activities of pomegranateCitation35. Urolithins A and B (8, 9) are the most abundant metabolites produced by human gut microbiota from dietary ellagic acid and ellagitannins, with urolithin A (8) being the most studied and common species found in natureCitation36–38. Although urolithin C (10) and D (11) are found to a lesser extent, specific pharmacological activities have also been reported for these gut microbiota metabolites. For instance, urolithin C (10) is able to elicit glucose-dependent activation of insulin secretion in pancreatic modelsCitation39. In another study, urolithin D (11) is identified as an inhibitor of EphA2/ephrin-A interaction with a competitive and reversible binding profile, supporting a putative mechanism for its colon cancer preventive propertiesCitation40. The scientific interest in the biological and pharmacological activities of urolithins (8–11) is further motivated by their bioavailability profile which is higher than ellagic acid (7), making such compounds good preclinical and clinical candidates for nutraceutical and drug discovery applicationsCitation37,Citation41,Citation42. To develop a structure-activity relationship (SAR) scheme for ellagic acid (7) and urolithins A–C (8–11) as ligands of Nsp5 and inhibitors of the SARS-CoV-2 viral cycle, we first investigated the physicochemical properties of these compounds. These properties could potentially affect the binding and inhibitory activities of these compounds towards the SARS-CoV-2 main protease Nsp5.
Physicochemical properties
The calculated octanol/water partition coefficient (cLogP) and the total polar surface area (TPSA) of compounds 7–11 were assessed using atom-based and fragment-based approaches, respectivelyCitation43,Citation44. The acidic dissociation constants (pKa) were both predicted and experimentally determined using a potentiometric titration method. In particular, the experimental pKa values of the phenolic groups of 7–11 were successfully determined when falling in a range between 2 and 12. The potentiometric method cannot provide reliable measurements outside this range of values due to the buffering reaction of waterCitation45. Hence, the very high predicted pKa values of two phenolic groups in ellagic acid (7) and urolithin D (11) as well as one phenolic group in urolithin C (10) were not determined with the potentiometric titration method (). reports the cLogP, TPSA, and experimental pKa values of compounds 7–11. Overall, a poor agreement is found between the predicted and the experimental pKa values of the compounds ( and ). Although ellagic acid (7) and urolithin D (11) are the most polar compounds for their highest number of phenolic groups, they differ in the relative pKa values. Ellagic acid has two experimentally determined pKa values, with the lowest value (pKa1 = 3.63) being likely ascribed to the hydrolysis of its lactone moieties (7-bis) occurring when the potentiometric titration reaches low pH values (pH < 6). The second pKa value (pKa2 = 7.04) can be assigned to either phenolic groups in position 3 and 3′ of 7, which thereby show identical or very similar pKa values that are not distinguished by the potentiometric method. Urolithin C (10) has three phenolic groups, yet only two of them have been experimentally determined with potentiometric titration (pKa1 = 6.58; pKa2 = 9.10). Urolithins A and B are di- and mono-hydroxylated metabolites of ellagic acid (7), respectively. The determination of their experimental pKa values shows that they are less acidic compounds than urolithins C and D.
Figure 4. Predicted pKa values of ellagic acid (7), its possible acid-hydrolysed product (7-bis), and urolithins A–D (8–11); predicted pKa values of phenolic groups are blue-labelled if they were not experimentally determinable; predicted pKa values are red-labelled if they were successfully determined by the potentiometric method ().
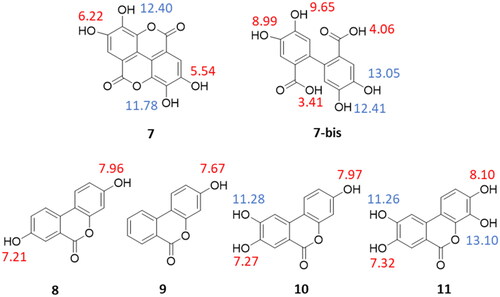
Table 1. Physicochemical properties of ellagic acid (7) and urolithins (8–11).
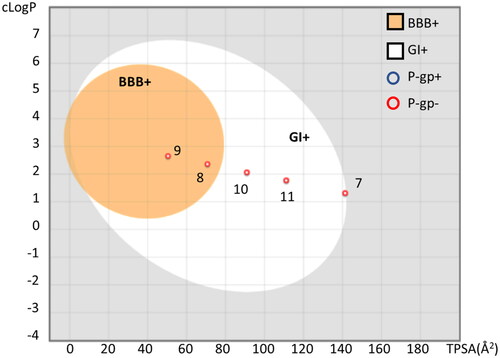
A putative bioavailability profile was next evaluated for each compound using the boiled-egg method that predicts gastro-intestinal (GI) absorption and the blood–brain barrier (BBB) permeability based on cLogP and TPSA valuesCitation46. Moreover, the likelihood of being P-gp substrate for ellagic acid (7) and urolithins (8–11) was also calculated using the SwissADME serverCitation47.
In agreement with the literature dataCitation37,Citation41,Citation42, results show that urolithins have better GI absorption than ellagic acid (). Moreover, urolithins B (9) and A (8) are also predicted as BBB permeant in view of their higher cLogP and lower TPSA values. It is worth noting that both ellagic acid (7) and urolithins A–D (8–11) are not predicted as P-gp substrates.
Figure 5. Boiled-egg representation of ellagic acid and urolithins A-C according to their TPSA and cLogP values, suggesting BBB permeability (BBB+, orange region), GI absorption (GI+, white region). Compounds predicted as P-gp substrate (P-gp+) are indicated with a blue circle; compounds predicted as negative P-gp substrate (P-gp−) are indicated with a red circle. The plot was generated using SwissADME.
Ligand binding activity
In this part of the study, we aimed to assess the interactions of ellagic acid (7) and urolithins A–D (8–11) to the recombinant SARS-CoV-2 main protease Nsp5. Ligand binding assays were performed using the MST methodologyCitation48. Hence, the recombinant SARS-CoV-2 main protease Nsp5 was labelled with a fluorescent dye (NT650-Nps5), and its stability and integrity were checked using nano-DSF assays, confirming that protein stability was not compromised after the time of incubation as well as structural integrity was preserved upon the labelling reaction (see Methods and Supplementary Materials, Figures S1, S2, Table S1). Buffer conditions were evaluated to identify the optimal set for MST signal reproducibility, using nirmatrelvir (1) as a positive control. Results show that all compounds are able to induce a variation of the thermophoretic movement of the ligand-bound NT650-Nps5 complex, generating binding curves (Supplementary Materials, Figures S3–S8) that are instrumental to calculate the relative dissociation constants (Kd) of the compounds ().
Table 2. Dissociation constants and Binding Efficiency Index (BEI) of nirmatrelvir (1), ellagic acid (7), and urolithins (8–11) to NT650-Nps5.
Overall, MST results are in agreement with the low micromolar dissociation constant reported for ellagic acid (7) in a recent SPR studyCitation28, yet they reveal distinct binding activities for urolithins A–D (8–11). Specifically, the inspection of reveals that ellagic acid (7) and urolithin D (11) are the most potent binders of the enzyme, with the Kd in the nanomolar range of potency. They are followed by urolithins C (10) and A (8) showing micromolar binding activities. Conversely, urolithin B (9) proves to be a much weaker binder of NT650-Nps5, yielding an incomplete binding curve at its maximum tested concentration (250 µM). These findings support a SAR scheme wherein a higher number of hydroxyl groups on the aromatic rings favours the inhibitory effect of these compounds against SARS-CoV-2 main protease Nsp5Citation28. Moreover, they also highlight the relevance of the pKa values. Indeed, the binding activity of the compounds seems to be influenced by the acidity of the molecule, with urolithin D (11) and ellagic acid (7) bearing a more acidic hydroxyl group that may account for their nanomolar potency.
Since ellagic acid (7) and urolithins (8–11) have a lower molecular weight than nirmatrelvir (1), the Binding Efficiency Index (BEI) was determined as a metric of the compound potency corrected for its sizeCitation49,Citation50. The BEI values reported in highlight the compounds that use their atoms most efficiently to engage SARS-CoV-2 main protease Nsp5 with molecular interactions, pinpointing those chemical structures that are more suitable to further medicinal chemistry optimisation efforts for improving their potency while maintaining good physicochemical properties. In this regard, it is worth noting that ellagic acid (7) and urolithins A (8), C (10), and D (11) show higher BEI values than nirmatrelvir (BEI = 16.8), with urolithin D being endowed with the most significant BEI value of 27.6.
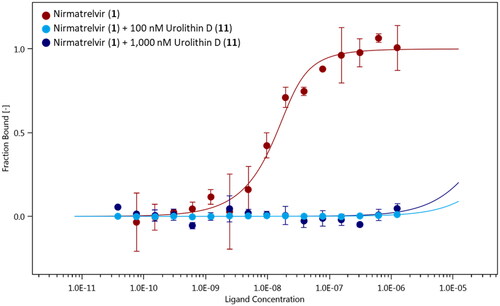
With the exception of urolithin A (8), the other active compounds (7, 10, and 11) bear a catechol group that has been listed as PAINS (Pan Assay INterference compoundS) motifCitation51. Catechol exhibits indeed different types of interference with assay readouts, including metal chelation, redox cycling, and unspecific reactivity with nucleophilic side chains of proteins. As a consequence, compounds bearing a catechol moiety are usually regarded as bad hits with poor evidence of structure–activity relationships and devoid of optimisation chances to high potent derivatives. Notwithstanding, the application of the PAINS concept to natural compounds is still a matter of debate, with the pivotal observation that many natural compounds contain such moieties accounting for their functional effectsCitation52. In this framework, a consensus view is that the PAINS concept may apply to natural products whenever they are found as low-micromolar potency hits and/or with unspecific reactivity in screening assays. Herein, urolithin D (11) was found as nanomolar potency hit in the MST assay. Hence, we next investigated the presence of a specific mechanism of reactivity to the catalytic site of SARS-CoV-2 main protease Nsp5, using a ligand binding displacement assay with nirmatrelvir (1). Specifically, the idea was to assess whether compound 11 could displace inhibitor 1 from the catalytic site of SARS-CoV-2 main protease Nsp5, leveraging its higher BEI and thereby suggesting a common binding site in the enzyme as well as a specific reactivity to the catalytic cysteine residue. In this regard, it is worth mentioning that nirmatrelvir (1) binds to the catalytic site of the enzyme, engaging Cys145 in the formation of a reversible covalent bond with its nitrile group (pdb codes: 7VH8, 7RFW, 7SI9, 7TE0)Citation14,Citation53–55. Ligand binding experiments of nirmatrelvir (1) to NT650-Nps5 were carried out in the presence of two increasing concentrations of urolithin D (100 nM and 1000 nM) to monitor the presence of a competitive binding behaviour with the rightward shift of the dissociation constant (Kd) of the inhibitor 1. It is found that urolithin D (11) abolishes the interaction of nirmatrelvir (1) to the enzyme, with the Kd value of the inhibitor 1 being not determinable in the presence of compound 11 at either of the tested concentrations ().
Figure 6. Binding curves of nirmatrelvir (1) to NT650-Nps5 in the absence (red points and line) and presence of 100 nM (light blue points and line) and 1000 nM (blue points and line) of urolithin D (11). Results are obtained from n0.3 independent measurements; bars represent the standard deviation of the points.
Molecular docking
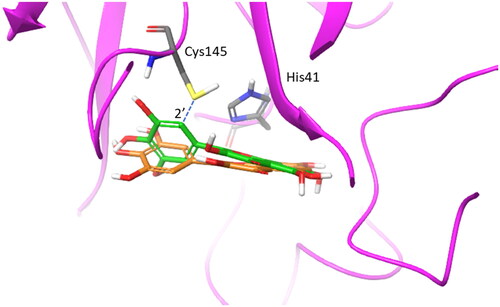
Having identified the binding site of urolithin D (11) within the catalytic cleft of SARS-CoV-2 main protease Nsp5, we next investigated the binding modes of ellagic acid (7) and urolithins (8–11) using molecular docking studies. In this regard, it is worth noting that these molecules belong to the chemical class of polyphenols as myricetin (2). Specifically, myricetin (2) has been reported as a potent irreversible inhibitor of Nsp5 (IC50 = 0.63 ± 0.01 μM), being able to engage Cys145 in a covalent bond with the aromatic carbon atom in 2′ position of its pyrogallol moiety (pdb code: 7B3E, 7DPP; )Citation16,Citation56.
Figure 7. Overlap of the experimental binding mode (green carbon sticks; pdb code 7B3E) and predicted binding mode (orange carbon sticks; top scored solution) of myricetin (2) as resulting from the docking study (RMSD = 1.57 Å). The RMSD is calculated between the corresponding heavy atoms of the flavonoid moiety of 2. The covalent bond between the aromatic carbon atom in 2′ position of 2 and Cys145 is shown as blue dashed line.
The reaction starts with an auto-oxidation process, which is common in polyphenols upon air exposureCitation57. In this process, the catechol moiety of 2 spontaneously oxidises with one hydroxyl group being firstly deprotonated to accelerate a transient formation of a o-semiquinone radical which secondly decays yielding a o-quinone moietyCitation58. This latter is a Michael acceptor group and reacts with the thiol group of Cys145, making a covalent adduct with the enzymeCitation56.
Docking studies of compounds 7–11 were thus carried out into the crystal structure of Nsp5 bound to myricetin (2) as detailed in the method section. The adopted docking algorithm (Glide) and the relative scoring function (Gscore) correctly identified the binding mode of myricetin (2) into the catalytic cleft with its carbon atom in 2′ position of the pyrogallol moiety laying in close proximity to Cys145 (; RMSD = 1.57 Å). Then, the top-scored binding mode for each compound was selected to assess the estimated binding energy (Gscore) and identify intermolecular interactions ().
Table 3. Docking scores (GScore, kcal/mol) of myricetin (2), ellagic acid (7), and urolithins (8–11) into the catalytic site of Nps5.
As a first result, the inspection of the binding energies (Gscore) reveals limitations of the scoring function to yield a correct estimation of these values since they are in poor agreement with the dissociation constants (Kd) of the docked compounds. Although this is generally known in literatureCitation59–61, the docking algorithm however succeeds in providing binding modes of myricetin (2) and compounds 7–11 that are compliant to react with the thiol group of Cys145.
To gain insights into structure-activity relationships, we then visually inspected the binding modes of compounds 7–11 into the catalytic site of Nsp5 (). All compounds show a conserved π-stacking interaction with the catalytic residue His41, whereas different number of hydrogen bonds are formed between the hydroxyl groups and the backbone or side chain polar atoms of residues shaping the catalytic cleft.
Figure 8. Binding mode (orange carbon sticks; top-scored solution) of ellagic acid (7) as resulting from the docking study. Catalytic site residues involved in hydrogen bond and π-stacking interactions () with 7 are bold labelled. Hydrogen bonds are shown as yellow dashed lines.
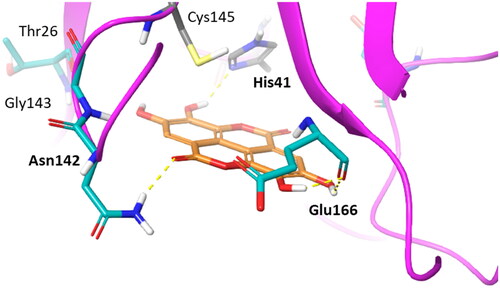
Figure 9. Binding modes (orange carbon sticks; top-scored solutions) of urolithins A–D (8, A; 9, B; 10, C; 11, D) as resulting from the docking study. Catalytic site residues involved in hydrogen bonds and π-stacking interactions with compounds 8–11 () are bold labelled. Hydrogen bonds are shown as yellow dashed lines.
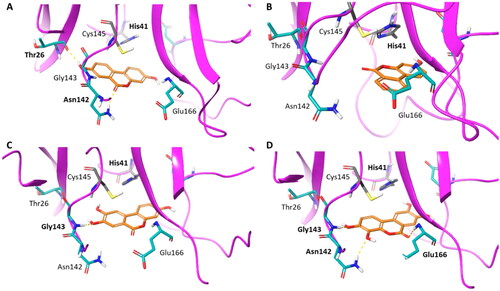
In agreement with the previous SAR schemeCitation28, compounds with the highest number of hydroxyl groups on the aromatic rings, namely ellagic acid (7) and urolithin D (11), show more hydrogen bond interactions with Nsp5 than other compounds (8–10; ), thereby accounting for their nanomolar binding potency. Moreover, the high number of hydroxyl groups favours an enhanced water solubility of compounds 7 and 11 that would be more available to bind to the enzyme.
However, this is not sufficient to explain the higher BEI of urolithins D (11; BEI = 27.6) and C (10; BEI = 21.2) than ellagic acid (7; BEI = 19.9) and urolithin A (8; BEI = 19.1). An explanation to such observation may rely on the different reactivity of these compounds for the formation of the Michael acceptor group. Indeed, the first step leading to the warhead is the deprotonation of one hydroxyl group of the catechol moiety for the generation of o-semiquinone radical. In this framework, it is worth noting that urolithins D and C show a lower pKa1 value than the pKa1 of urolithin A and pKa2 of ellagic acid, corresponding to the acidic hydrogen of the hydroxyl group of the catechol moiety. Hence, the lower pKa value of urolithins D and C may increase compound’s reactivity, favouring the warhead formation.
Cell-based antiviral activity
The most potent and promising compound, namely urolithin D, was submitted to cellular experiments using Vero E6 cells with the aim of assessing its cytotoxicity as well as its antiviral activity. Accordingly, urolithin D (11) and nirmatrelvir (1) were used in preliminary experiments to assess the effect on Vero E6 cell viability. To this end, the cells were incubated with different concentrations of each compound for 24 h, and cell viability was assessed by MTT reduction assay. As shown in , neither urolithin D (11) or nirmatrelvir (1) were able to significantly affect cell metabolic activity, assumed to be well-tolerated by Vero E6 cells at all the concentrations tested.
Figure 10. Cytotoxic effect of urolithin D (11) and nirmatrelvir (1) on Vero E6 cells. Cells were incubated with different concentrations of urolithin D (A) or nirmatrelvir (B) for 24 h and cellular viability was assessed through the MTT reduction assay. Data are expressed as mean ± SD of cell viability percentage with respect to control from two independent experiments performed in triplicate.
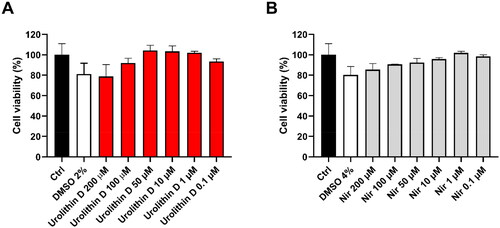
Of note, the apparent slight reduction of cell viability by the highest concentrations of compounds could be ascribed to the corresponding amount of DMSO used to dissolve urolithin D (11) and nirmatrelvir (1). Consequently, the subsequent experiments were performed using 50 µM of urolithin D (11) and nirmatrelvir (1) as the highest concentration.
The antiviral activity of urolithin D (11) was assessed by a Vero E6 cell-based assay. The cells were infected with the SARS-CoV-2 BA.5 strain and treated with different concentrations of compound 11 and nirmatrelvir (1) as the positive control. After incubations, the supernatant viral titre was determined through the plaque assay. As shown in , the viral replication was not affected by the treatment with urolithin D (11) at all the tested concentrations. Indeed, the viral titres of the supernatants were completely comparable to the untreated control. Conversely, the protease inhibitor nirmatrelvir (1) was able to significantly reduce the number of infective particles recovered from the treated samples ().
Figure 11. Antiviral activity of urolithin D (11) and nirmatrelvir (1). Cells were infected with 0.01 multiplicity of infection of SARS-CoV-2 BA.5 variant and treated with different concentrations of (A) urolithin D or (B) nirmatrelvir for 48 h. After incubation, the supernatant viral load was assessed through plaque assay. Viral titres are expressed as mean ± SD of plaque-forming units (PFU)/mL from two experiments performed in triplicate. *p < 0.05, antiviral-treated sample vs. vehicle-treated sample.
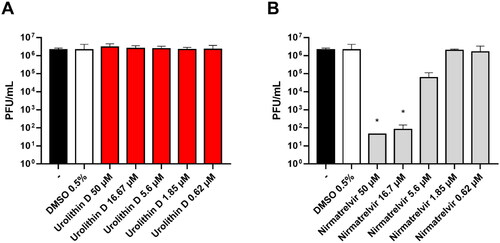
These results are in contrast to a previous work which supports the antiviral activity of polyphenolic compounds in Vero E6 cells, reporting the good inhibition activity of myricetin (2, EC50 = 8.0 µM) against the replication of SARS-CoV-2Citation56. In another study, however, myricetin was found devoid of antiviral SARS-CoV-2 activity using a cytopathic effect (CPE) inhibition assay in Vero E6 cellsCitation16. In the latter case, the authors proposed that the longer cellular readout (72 h) of the experiment favoured metabolic and/or efflux processes of myricetin (2) that could lead to diminishing the intracellular concentration of the polyphenol below an efficiency threshold to achieve inhibition of viral replication. This observation may also tentatively explain the lack of antiviral activity for urolithin D (11) in our cellular study, wherein a similarly long time of readout was used for the appraisal of the inhibition of SARS-CoV-2 replication.
Other studies have also pointed out the hypothesis that treating Vero E6 cells with flavonoids first and then virus infection, as in the experiments of Su et al.Citation56, may highlight a preventive role of this class of compounds through their interference with other mechanisms of viral infection, such as the interaction of the spike protein with the ACE2 receptor. Citation62,Citation63
Conclusions
Ellagic acid (7) and its gut microbiota metabolites, urolithins A–D (8–11), are natural polyphenols endowed with distinct biological activitiesCitation32,Citation33. In this study, we first investigated the physicochemical properties and binding activities of these compounds against the main protease Nsp5 of SARS-CoV-2. In agreement with the literature data, urolithins A–D (8–11) are predicted with a better bioavailability profile than ellagic acid (7). MST binding experiments show that ellagic acid (7) and urolithins A–D (8–11) are able to bind Nsp5 with dissociation constants (Kd) ranging from high micromolar (urolithin B, 9) to nanomolar potencies (urolithin D, 11). The inspection of the Binding Efficiency Index highlights urolithin D (11, BEI = 27.6) as the compound that exploits its atoms most efficiently to engage the main protease Nsp5 with molecular interactions. Noteworthy, the BEI of compound 11 is also significantly higher than nirmatrelvir (1, BEI = 16.8). Ligand binding displacement assays amid urolithin D (11) and nirmatrelvir (1) reveal a competitive binding mode to Nsp5 of SARS-CoV-2, pinpointing the catalytic cleft as the binding site of 11 as well as its specific reactivity to Cys145. This observation is consistent with crystallisation studies of its polyphenol analogue, myricetin (2), which has been reported as an irreversible inhibitor of Nsp5 that engages the catalytic Cys145 in a covalent bond. Hence, ellagic acid (7) and urolithins A–D (8–11) may work as irreversible inhibitors of Nsp5, sharing key conserved interactions with Cys145 and His41 in the catalytic cleft as resulting from docking studies. Of note, the lower pKa1 value of the catechol hydroxylic group of urolithin D (11) is a pivotal physicochemical property that may account for the nanomolar potency and high BEI of this compound. Indeed, the more acidic proton favours a higher reactivity of compound 11 for the formation of the Michael acceptor group to readily engage Cys145 in a covalent bond.
Unfortunately, these promising results have not successfully translated into cellular data. At odds with nirmatrelvir (1), indeed, it is found that urolithin D (11) is devoid of antiviral activity against SARS-CoV-2 BA.5 strain in a Vero E6 cell-based assay. In this regard, however, it should be mentioned that contradictory results were also reported for the antiviral SARS-CoV-2 activity of its polyphenol analogue, myricetin (2) when using different experimental protocols of inhibition of viral replication in Vero E6 cellsCitation16,Citation56. Since this class of compounds has been reported as accounting for the antiviral activity of some functional foodsCitation32,Citation33, it is our opinion that further work is required to investigate the effects of different experimental protocols of cellular assays, including time of readout, on the antiviral activity of polyphenols. These studies will provide insight into how customising experimental protocols can affect the clinical translatability of preclinical studies, enabling a more comprehensive evaluation of the potential of polyphenols, such as ellagic acid and urolithins, for nutraceutical and/or drug discovery applications. Last but not least, the high binding potency observed for these natural products, as well as the presence of structure-activity relationships and a specific reactivity to Cys145 suggest that they can be considered more than merely PAINS in the context of viral protease inhibitors, thereby deserving further investigations.
Methods
Physicochemical property profiling
Predictions of the acidic constants were obtained with the calculator plugin Marvin (v23.5, 2023; ChemAxon; www.chemaxon.com). Experimental values of the acidic constants (pKa) were determined at 25 °C using Sirius T3 potentiometric method (Pion Inc, E. Sussex, UK)Citation45. To prevent solubility issues during the dissolution stage in Ionic Strength Adjusted (ISA) water (KCl 0.15 M), compounds 7–11 were dissolved in 100% DMSO at the concentration of 50 mM. All the experiments were performed specifying the sample volume, number of expected pKa values, and their predicted values according to the calculator plugin Marvin. Three titrations were performed for each experiment to measure the pKa of the relative compound. The titration mode was set starting from high pH (pH = 12) to reach low pH (pH = 2). The Sirius T3 control software v1.1.3.0 was instrumental to control the titration experiment. Titrations were performed at different methanol/water ratios (50% methanol, 40% methanol, and 30% methanol), using the Yasuda-Shedlovsky method to extrapolate the relative aqueous pKa values of the compoundsCitation64. Results were analysed using the Sirius T3 refine software v1.1.3.0. The reliability of the titration assays was assessed by inspecting the R2 resulting from the Yasuda-Shedlovsky method. Results were deemed acceptable if R2 > 0.9. The accuracy of the titration assays was evaluated by inspecting the buffering capacity signal of the analyte which should be above the intrinsic buffering capacity of water for an accurate potentiometric measurement. The experimental pKa values of phenolic groups of compounds 7–11 were successfully determined when falling in a range between 2 and 12.
The octanol/water partition coefficient (cLogP), the total polar surface area (TPSA), and the likelihood of being P-gp substrates of compounds 7–11 were calculated using the SwissADME serverCitation47. These molecular descriptors were instrumental to assess the gastro-intestinal (GI) absorption and the blood–brain barrier (BBB) permeability of the compounds using the boiled-egg methodCitation46.
Protein production
The expression of the full-length SARS-CoV-2 NSP5 was performed by transforming E. Coli BL21 DE3 cells with a PGEX-6P-1 plasmid containing the gene sequence of the ORF1ab polyprotein residues 3264–3569 with an additional N-terminal AVLQ and C-terminal GPHHHHHH. This gene sequence has been optimised for E. Coli expression and cloned between the BamHI and NotI restriction sites of plasmid pGEX-6P-1, as previously reportedCitation65.
The construct is designed as the NSP5 protein fused at the N-terminal with the GST protein, followed by the NSP5 recognition site such that auto-cleavage of NSP5 itself occurs in the cell during gene expression. The C terminus is fused to the 3C pre-scission protease sequence site followed by His tag suitable for affinity purification. Cells were resuspended in 80 ml of buffer 1 (20 mM Tris, 150 mM NaCl, pH 7.8) sonicated for 30 min (2 s on and 10 off) and ultra centrifugated at 40 000 rpm for 40 min. The supernatant was loaded on a His-trap FF HP column. The column was washed with 150 ml of buffer 1 to remove unbound proteins before NSP5 was eluted against a linear gradient of 20 column volumes (CV) from 0 to 500 mM imidazole concentration.
The fractions containing the protein were collected, then 1 mg of 3 C precision protease was added and the solution was dialysed against 2 L of buffer 2 (20 mM Tris, 150 mM NaCl, 1 mM DTT, pH 7.8). To separate the non-cleaved NSP5-His tagged and the GST-tagged 3 C pre-scission the solution was passed through a His-Trap HP FF and a GST-Trap HP FF columns. A final purification step was carried out by injecting the protein into a gel filtration column (Superdex 200 Increase 16/60 75 pg) previously equilibrated with buffer 3 (20 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH 7.8).
Stability and integrity checks of the protein
The stability and integrity of the recombinant Nsp5 protein were evaluated using nano-Differential Scanning Fluorimetry (nano-DSF) assays. In particular, the stability of the protein was checked after 48 h, with the protein kept at +4 °C in the dark, in a buffer pH 8. The integrity of the protein was checked upon the labelling reaction with RED dye NT650 (Monolith Protein Labelling Kit RED-NHS 2nd generation; NanoTemper Technologies, Munich, Germany) using a 1:3 ratio (protein:dye). The labelling reaction was performed mixing a volume of 100 μL of 28 μM solution of recombinant Nsp5 with 100 μL of 84 μM RED dye NT650 in the labelling buffer (130 mM NaHCO3, 50 mM NaCl, pH 8.2) supplied by NanoTemper. The labelling solution was incubated for 30 min and the unreached fluorophore was removed using a size-exclusion chromatography column, equilibrated with 20 mM TRIS, 140 mM NaCl, pH 8.0 (TRIS buffer). Nano-DSF experiments were run with Tycho NT.6 instrument (NanoTemper Technologies, Munich, Germany) monitoring protein thermal denaturation by variations of the intrinsic fluorescence emission of tryptophane and tyrosine residues at 330 and 350 nm during the heating phaseCitation65. A solution of 3 µM Nsp5 was prepared in TRIS buffer for such experiments. The assays were executed in triplicate and the inflection Temperature (Ti) was calculated by the Tycho NT.6 software (v1.3.2.880). Results are reported in the Supplementary Materials (Figures S1, S2 and Table S1). As a result, no variation was found amid the different Nsp5 samples (ΔTi < 1.0), confirming protein stability after 48 h at +4 °C buffer pH 8 in the dark as well as protein integrity upon the labelling process.
Ligand binding assays
Thermophoresis is the directional movement of a biomolecular complex along a temperature gradient. The movement is dependent on the size, charge, and hydration shell of the complex that changes upon a binding event between a target protein and its cognate ligandCitation66. In a typical MST assay, a fluorescently labelled target protein is kept at constant concentration while it is titrated with serial dilutions of the unlabelled cognate ligand. An IR-laser induces a temperature gradient and the protein-bound fluorophores in the solution are excited. The migration of the biomolecular complex occurs typically from the hot to the cold region, inducing a change in the fluorescence signal. This variation is expressed as the normalised fluorescence (Fnorm) and used to produce a binding curve for deriving the dissociation constant (Kd) between the target protein and the cognate ligandCitation48.
The recombinant Nsp5 protein was fluorescently labelled with RED dye NT650, using a 1:3 ratio (protein:dye). Protein and dye concentrations were determined by absorption spectroscopy using a Thermo Scientific™ NanoDrop™ One spectrophotometer (Thermo Fisher Scientific Inc., Waltham Massachusetts, USA) and applying EquationEquations (1)(1)
(1) and Equation(2)
(2)
(2) , respectively. In the equations, the extinction coefficients are ɛprotein = 32 890 M−1 cm−1 at 280 nm for recombinant Nps5, εRED dye = 195 000 M−1 cm−1 at 650 nm for NT650 fluorophore, with a correction factor of 0.04. The degree of labelling (DOL) was calculated using EquationEquation (3)
(3)
(3) , yielding a value of 0.5 for NT650-Nsp5
(1)
(1)
(2)
(2)
(3)
(3)
Ligand binding experiments of compounds 1 (MedChemExpress, Cat. No. HY-138687), 7–11 (Sigma-Aldrich, Cat. No. E2250, SML1791, SML1649, SML3047, and SMB01339, respectively) were performed in TRIS buffer, adding 0.01% Tween20, 1 mM EDTA and 2% DMSO. Each compound was tested against NT650-Nps5 (protein concentration 50 nM) at buffer pH 8 with sixteen serial dilutions starting from a concentration of 1 µM for nirmatrelvir (1); 250 µM for ellagic acid (7), urolithins A (8) and B (9); 1 mM for urolithin C (10); 30 µM for urolithin D (11). After overnight incubation at +4 °C in the dark, samples were loaded into 16 premium coated capillary tubes (MO − K025; NanoTemper Technologies, Munich, Germany) for the MST analysis. MST signals were recorded for each capillary at medium laser power, and 40% LED power, and analysed in initial fluorescence mode. The Binding Efficiency Index (BEI)Citation67 was calculated using the following EquationEquation (4)(4)
(4) :
(4)
(4)
The location of the binding site for the most potent compound, namely urolithin D (11), was investigated using a ligand displacement assay with MST and nirmatrelvir (1) as reference compounds. In this experiment, NT650-Nps5 (protein concentration 20 nM) was incubated with two concentrations of compound 11 (100 and 1000 nM) overnight at +4 °C and buffer pH 8 in the dark, before adding nirmatrelvir (1) with sixteen serial dilutions starting from a concentration of 1 µM. MST signals were recovered using 50% LED power, medium MST power, and setting the hot region in the Manual mode between 19/20s.
All MST experiments were run in triplicate and resulting data were processed using MO.Affinity Analysis v2.3 (NanoTemper Technologies, Munich, Germany). Dissociation constants (Kd) were obtained as mean values with a confidence interval (±) that defines the range where the Kd falls with a 68% of certainty, as declared by NanoTemper. The signal to noise ratio (S/N) was >12 in all MST experiments, indicating excellent assay conditions. MST experiments were carried out using Monolith NT.115 instrument (NanoTemper Technologies, Munich, Germany).
Molecular docking calculations
The crystal structure of Nsp5 bound to myricetin (2) with the highest resolution factor (pdb code: 7B3E; resolution 1.77 Å)Citation16 was used for the docking studies of myricetin (2), ellagic acid (7), and urolithins A–D (8–11). The chemical structures of these compounds were taken from the PubChem databaseCitation68 and refined using LigPrep (Schrödinger Release 2021–3; LigPrep, Schrödinger, LLC, New York, NY, USA, 2021) to add hydrogens, to generate all ionisation and tautomeric states at pH = 7 ± 2, and to optimise their internal geometry with the OPLS4 force field until reaching an energy gradient convergence criterion of 0.05 kJ/Å-mol. The atomic coordinates of 7B3E chain A were kept, removing the co-crystallized ligand (2) bound to Cys145 of the catalytic cavity. The Protein Preparation Wizard (PPW) tool implemented in Maestro (Schrödinger Release 2021–3; Maestro, Schrödinger, LLC, New York, NY, 2021) was used to process and energetically refine 7B3E chain A, removing water and any detergent molecule, adding hydrogen atoms and optimising internal geometries. The OPLS4 force field was used with an atomic coordinate displacement restrain on heavy atoms set to 0.30 Å. Docking studies of myricetin (2), ellagic acid (7), and urolithins A–D (8–11) into the catalytic site of 7B3E chain A were performed with Glide (Schrödinger Release 2021–3: Glide, Schrödinger, LLC, New York, NY, USA, 2021)Citation69. The grid box was defined with its centre located close to Cys145 (x = 20.83, y = −0.08, z = −23.36). The inner grid box was sized 10 × 10 × 10 Å. Docking studies were carried out using the standard precision (SP) method and the GScore scoring function (kcal/mol). Criteria for selection of the initial pose were set to keep n 50 000 poses per ligand for the initial phase of docking, to use a scoring window of 500 kcal/mol, to keep the best n 1000 poses per ligand for energy minimisation, to use the expanded sampling modeCitation60. Six independent docking calculations were run for each compound. It was found that all of them show an identical conserved binding mode in the top-scored pose with a very low standard deviation of the relative GScore among docking replicas (Table S2 of Supplementary Materials).
The good performance of this docking protocol was also validated in reproducing the experimental binding mode of myricetin (2) into the catalytic cleft of 7B3E chain A, as evidenced by the low root mean square deviation (RMSD) among heavy atoms of the top-scored pose and the corresponding atoms of the experimental pose of compound 2 (, RMSD = 1.57 Å). Hence, the top-scored pose of each compound was stored for visual inspection and generation of .
Cell-based antiviral activity assay
SARS-CoV-2 BA.5 (also known as the Omicron 5 variant) strain was obtained from a nasopharyngeal swab of a symptomatic patient of “Santa Maria della Misericordia” Hospital of Perugia, Perugia, Italy, as previously describedCitation70. Frozen viral stocks titered by Half-maximal Tissue Culture Infectious Dose (TCID50) endpoint dilution assayCitation71 were kept at −80 °C and thawed immediately before each experiment. Whole genome sequencing for variant assignation was performed as previously describedCitation72,Citation73. Vero E6 ATCC CRL-1586 cells were maintained in Eagle’s minimum essential medium (MEM) containing 10% foetal bovine serum (FBS) and 1% (v/v) penicillin–streptomycin at 37 °C with 5% CO2. The Vero E6 cell line was kindly provided by Istituto Zooprofilattico Sperimentale di Brescia, Brescia, Italy.
Nirmatrelvir (PF-07321332, 1) was purchased from MedChemExpress (Monmouth Junction, NJ, USA) and dissolved in DMSO according to the manufacturer’s instructions. Small aliquots were then kept at −80 °C, thawed before each experiment, and discarded after use. The cytotoxic effect of selected compounds was assessed by the MTT [3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] reduction assay. MTT was purchased from Merck (Merck, Darmstadt, Germany), dissolved in sterile PBS at the concentration of 5 mg/mL, and filter-sterilized immediately before use. Vero E6 cells (100 µL, 1 × 105/mL) were dispensed into each well of a 96-well plate and incubated at 37 °C with 5% CO2 for 24 h. After incubation, the medium was discarded, and the monolayers were incubated with 100 µL of urolithin D (11) or nirmatrelvir (1) (200–0.1 µM) for another 24 h in the same conditions. Simple medium and medium with different concentrations of DMSO (4, 2, 1, 0.5, and 0.1%) were used as control. After treatments, MTT stock solution was diluted 1:10 with complete medium and 100 µL was added to each well of the plate and subsequently incubated for 3 h in the same conditions. Finally, 100 µL of DMSO was used to dissolve formazan crystals precipitated on the bottom of the wells after incubation for 1 h. Absorbance values at 570 nm (reference filter at 630 nm) were obtained using a microplate reader (Tecan Infinite M200, Tecan Trading AG, City, Switzerland). The percentages of cytotoxicity of each antiviral were calculated based on cells treated with the respective vehicle concentrations (medium with DMSO).
Anti-SARS-CoV-2 activity of compounds was tested as previously describedCitation74,Citation75. Vero E6 cells (3 × 104 cells/well) were cultured in 96-well plates at 37 °C with 5% CO2 for 24 h. After incubation, cells were infected with a multiplicity of infection (MOI) of 0.01. SARS-CoV-2 BA.5 strain was allowed to adsorb to the surface of cells for 1 h at 37 °C. Then, the virus inoculum was removed, and cells were treated with 3-fold serial dilutions (50–0.62 µM) of urolithin D (11) and nirmatrelvir (1). In each plate, negative controls (complete medium and medium with 0.5, 0.25, and 0.1% of DMSO) and infected positive controls (SARS-CoV-2 alone) were included. Plates were incubated at 37 °C with 5% CO2 for 48 h. After incubation, cell supernatants were stored at −80 °C for subsequent analysis.
The cell supernatants obtained in previous experiments were used for virus titre determination with plaque reduction assay, as previously describedCitation75. Vero E6 cells (3 × 105 cells/well) were cultured in 12-well plates for 24 h at 37 °C with 5% CO2. The medium of each well was replaced with 250 µL of 10-fold serial dilution of supernatants and incubated for 1 h, rocking the plate by hand every 15 min. The supernatant dilutions were removed and 1 ml of overlay medium (complete medium with agar 0.1%) was added to each well for 72 h. After that, the cells were fixed and stained with a solution containing 4% formalin and 0.5% crystal violet. Viral titre was expressed as plaque-forming units (PFU)/mL, considering wells with 2–50 plaques.
Statistical analysis was performed using Prism Graphpad v.8.3 (GraphPad Software, San Diego, CA, USA). Data were tested for normality using the Kolmogorov–Smirnov test and were presented as mean with the respective standard deviation (SD) or median with interquartile range (IQR), as appropriate. A p-values <0.05 was considered as significant.
Supplemental Material
Download PDF (705.4 KB)Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Galea S, Abdalla SM. COVID-19 pandemic, unemployment, and civil unrest: underlying deep racial and socioeconomic divides. JAMA. 2020;324(3):227–228.
- Rudrapal M, Khairnar SJ, Borse LB, Jadhav AG. Coronavirus disease-2019 (COVID-19): an updated review. Drug Res. 2020;70(9):389–400.
- Paul E. Address in PATHOLOGY ON CHEMOTHERAPEUTICS. Lancet. 1913;182(4694):445–451.
- de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523–534.
- Muramatsu T, Takemoto C, Kim Y-T, Wang H, Nishii W, Terada T, Shirouzu M, Yokoyama S. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc Natl Acad Sci USA. 2016;113(46):12997–13002.
- Yang H, Xie W, Xue X, Yang K, Ma J, Liang W, Zhao Q, Zhou Z, Pei D, Ziebuhr J, et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLOS Biol. 2005;3(10):e324.
- Yang H, Bartlam M, Rao Z. Drug design targeting the main protease, the achilles heel of coronaviruses. Curr Pharm Des. 2006;12(35):4573–4590.
- Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412.
- Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293.
- Silvestrini L, Belhaj N, Comez L, Gerelli Y, Lauria A, Libera V, Mariani P, Marzullo P, Ortore MG, Palumbo Piccionello A, et al. The dimer-monomer equilibrium of SARS-CoV-2 main protease is affected by small molecule inhibitors. Sci Rep. 2021;11(1):9283.
- Paciaroni A, Libera V, Ripanti F, Orecchini A, Petrillo C, Francisci D, Schiaroli E, Sabbatini S, Gidari A, Bianconi E, et al. Stabilization of the dimeric state of SARS-CoV-2 main protease by GC376 and nirmatrelvir. Int J Mol Sci. 2023;24(7):6062.
- Liu Y, Liang C, Xin L, Ren X, Tian L, Ju X, Li H, Wang Y, Zhao Q, Liu H, et al. The development of coronavirus 3C-Like protease (3CLpro) inhibitors from 2010 to 2020. Eur J Med Chem. 2020;206:112711.
- Pillaiyar T, Manickam M, Namasivayam V, Hayashi Y, Jung SH. An overview of severe acute respiratory syndrome–coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J Med Chem. 2016;59(14):6595–6628.
- Owen DR, Allerton CMN, Anderson AS, Aschenbrenner L, Avery M, Berritt S, Boras B, Cardin RD, Carlo A, Coffman KJ, et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374(6575):1586–1593.
- Gidari A, Sabbatini S, Schiaroli E, Bastianelli S, Pierucci S, Busti C, Comez L, Libera V, Macchiarulo A, Paciaroni A, et al. The combination of molnupiravir with nirmatrelvir or GC376 has a synergic role in the inhibition of SARS-CoV-2 replication in vitro. Microorganisms. 2022;10(7):1475.
- Kuzikov M, Costanzi E, Reinshagen J, Esposito F, Vangeel L, Wolf M, Ellinger B, Claussen C, Geisslinger G, Corona A, et al. Identification of inhibitors of SARS-CoV-2 3CL-pro enzymatic activity using a small molecule in vitro repurposing screen. ACS Pharmacol Transl Sci. 2021;4(3):1096–1110.
- Günther S, Reinke PYA, Fernández-García Y, Lieske J, Lane TJ, Ginn HM, Koua FHM, Ehrt C, Ewert W, Oberthuer D, et al. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372(6542):642–646.
- Mani JS, Johnson JB, Steel JC, Broszczak DA, Neilsen PM, Walsh KB, Naiker M. Natural product-derived phytochemicals as potential agents against coronaviruses: a review. Virus Res. 2020;284:197989.
- Nguyen TTH, Woo H-J, Kang H-K, Nguyen VD, Kim Y-M, Kim D-W, Ahn S-A, Xia Y, Kim D. Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol Lett. 2012;34(5):831–838.
- Jo S, Kim S, Shin DH, Kim MS. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem. 2020;35(1):145–151.
- Jo S, Kim H, Kim S, Shin DH, Kim MS. Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem Biol Drug Des. 2019;94(6):2023–2030.
- Park J-Y, Ko J-A, Kim DW, Kim YM, Kwon H-J, Jeong HJ, Kim CY, Park KH, Lee WS, Ryu YB, et al. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J Enzyme Inhib Med Chem. 2016;31(1):23–30.
- Chen L-R, Wang Y-C, Lin YW, Chou S-Y, Chen S-F, Liu LT, Wu Y-T, Kuo C-J, Chen TS-S, Juang S-H, et al. Synthesis and evaluation of isatin derivatives as effective SARS coronavirus 3CL protease inhibitors. Bioorg Med Chem Lett. 2005;15(12):3058–3062.
- Liu W, Zhu H-M, Niu G-J, Shi E-Z, Chen J, Sun B, Chen W-Q, Zhou H-G, Yang C. Synthesis, modification and docking studies of 5-sulfonyl isatin derivatives as SARS-CoV 3C-like protease inhibitors. Bioorg Med Chem. 2014;22(1):292–302.
- Ryu YB, Jeong HJ, Kim JH, Kim YM, Park J-Y, Kim D, Nguyen TTH, Park S-J, Chang JS, Park KH, et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg Med Chem. 2010;18(22):7940–7947.
- Park J-Y, Kim JH, Kim YM, Jeong HJ, Kim DW, Park KH, Kwon H-J, Park S-J, Lee WS, Ryu YB, et al. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg Med Chem. 2012;20(19):5928–5935.
- Khalifa I, Zhu W, Mohammed HHH, Dutta K, Li C. Tannins inhibit SARS‐CoV‐2 through binding with catalytic dyad residues of 3CLpro : an in silico approach with 19 structural different hydrolysable tannins. J Food Biochem. 2020;44(10):e13432.
- Li H, Xu F, Liu C, Cai A, Dain JA, Li D, Seeram NP, Cho BP, Ma H. Inhibitory effects and surface plasmon resonance-based binding affinities of dietary hydrolyzable tannins and their gut microbial metabolites on SARS-CoV-2 main protease. J Agric Food Chem. 2021;69(41):12197–12208.
- Adelusi TI, Oyedele A-QK, Monday OE, Boyenle ID, Idris MO, Ogunlana AT, Ayoola AM, Fatoki JO, Kolawole OE, David KB, et al. Dietary polyphenols mitigate SARS-CoV-2 main protease (Mpro)–molecular dynamics, molecular mechanics, and density functional theory investigations. J Mol Struct. 2022;1250:131879.
- Rudrapal M, Issahaku AR, Agoni C, Bendale AR, Nagar A, Soliman MES, Lokwani D. In silico screening of phytopolyphenolics for the identification of bioactive compounds as novel protease inhibitors effective against SARS-CoV-2. J Biomol Struct Dyn. 2022;40(20):10437–10453.
- Rudrapal M, Celik I, Chinnam S, Azam Ansari M, Khan J, Alghamdi S, Almehmadi M, Zothantluanga JH, Khairnar SJ. Phytocompounds as potential inhibitors of SARS-CoV-2 Mpro and PLpro through computational studies. Saudi J Biol Sci. 2022;29(5):3456–3465.
- Landete JM. Ellagitannins, ellagic acid and their derived metabolites: a review about source, metabolism, functions and health. Food Res Int. 2011;44(5):1150–1160.
- Evtyugin DD, Magina S, Evtuguin DV. Recent advances in the production and applications of ellagic acid and its derivatives. a review. Molecules. 2020;25(12):2745.
- Ríos JL, Giner RM, Marín M, Recio MC. A pharmacological update of ellagic acid. Planta Med. 2018;84(15):1068–1093.
- Howell AB, D'Souza DH. The pomegranate: effects on bacteria and viruses that influence human health. Evid Based Complement Alternat Med. 2013;2013:1–11.
- Espín JC, Larrosa M, García-Conesa MT, Tomás-Barberán F. Biological significance of urolithins, the gut microbial ellagic acid-derived metabolites: the evidence so far. Evid Based Complement Alternat Med. 2013;2013:1–15.
- Tomás-Barberán FA, González-Sarrías A, García-Villalba R, Núñez-Sánchez MA, Selma MV, García-Conesa MT, Espín JC. Urolithins, the rescue of “old” metabolites to understand a “new” concept: metabotypes as a nexus among phenolic metabolism, microbiota dysbiosis, and host health status. Mol Nutr Food Res. 2017;61(1):1500901.
- D'Amico D, Andreux PA, Valdés P, Singh A, Rinsch C, Auwerx J. Impact of the natural compound urolithin A on health, disease, and aging. Trends Mol Med. 2021;27(7):687–699.
- Bayle M, Neasta J, Dall’Asta M, Gautheron G, Virsolvy A, Quignard J-F, Youl E, Magous R, Guichou J-F, Crozier A, et al. The ellagitannin metabolite urolithin C is a glucose‐dependent regulator of insulin secretion through activation of L‐type calcium channels. Br J Pharmacol. 2019;176(20):4065–4078.
- Giorgio C, Mena P, Del Rio D, Brighenti F, Barocelli E, Hassan-Mohamed I, Callegari D, Lodola A, Tognolini M. The ellagitannin colonic metabolite urolithin D selectively inhibits EphA2 phosphorylation in prostate cancer cells. Mol Nutr Food Res. 2015;59(11):2155–2167.
- González-Barrio R, Borges G, Mullen W, Crozier A. Bioavailability of anthocyanins and ellagitannins following consumption of raspberries by healthy humans and subjects with an ileostomy. J Agric Food Chem. 2010;58(7):3933–3939.
- Cerdá B, Espín JC, Parra S, Martínez P, Tomás-Barberán FA. The potent in vitro antioxidant ellagitannins from pomegranate juice are metabolised into bioavailable but poor antioxidant hydroxy-6H-dibenzopyran-6-one derivatives by the colonic microflora of healthy humans. Eur J Nutr. 2004;43(4):205–220.
- Wildman SA, Crippen GM. Prediction of physicochemical parameters by atomic contributions. J Chem Inf Comput Sci. 1999;39(5):868–873.
- Ertl P, Rohde B, Selzer P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J Med Chem. 2000;43(20):3714–3717.
- Dardonville C. Automated techniques in pKa determination: low, medium and high-throughput screening methods. Drug Discov Today Technol. 2018;27:49–58.
- Daina A, Zoete V. A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem. 2016;11(11):1117–1121.
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7(1):42717.
- Jerabek-Willemsen M, André T, Wanner R, Roth HM, Duhr S, Baaske P, Breitsprecher D. Microscale thermophoresis: interaction analysis and beyond. J Mol Struct. 2014;1077:101–113.
- Schultes S, De Graaf C, Haaksma EEJ, De Esch IJP, Leurs R, Krämer O. Ligand efficiency as a guide in fragment hit selection and optimization. Drug Discov Today Technol. 2010;7(3):e157–e162.
- Abad-Zapatero C, Metz JT. Ligand efficiency indices as guideposts for drug discovery. Drug Discov Today. 2005;10(7):464–469.
- Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53(7):2719–2740.
- Baell JB. Feeling nature’s PAINS: natural products, natural product drugs, and pan assay interference compounds (PAINS). J Nat Prod. 2016;79(3):616–628.
- Zhao Y, Fang C, Zhang Q, Zhang R, Zhao X, Duan Y, Wang H, Zhu Y, Feng L, Zhao J, et al. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell. 2022;13(9):689–693.
- Kneller DW, Li H, Phillips G, Weiss KL, Zhang Q, Arnould MA, Jonsson CB, Surendranathan S, Parvathareddy J, Blakeley MP, et al. Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease. Nat Commun. 2022;13(1):2268.
- Yang KS, Leeuwon SZ, Xu S, Liu WR. Evolutionary and structural insights about potential SARS-CoV-2 evasion of nirmatrelvir. J Med Chem. 2022;65(13):8686–8698.
- Su H, Yao S, Zhao W, Zhang Y, Liu J, Shao Q, Wang Q, Li M, Xie H, Shang W, et al. Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease. Nat Commun. 2021;12(1):3623.
- Yang J, Cohen Stuart MA, Kamperman M. Jack of all trades: versatile catechol crosslinking mechanisms. Chem Soc Rev. 2014;43(24):8271–8298.
- Doona CJ, Kustin K. Kinetics and mechanism of pyrogallol autoxidation: Calibration of the dynamic response of an oxygen electrode. Int J Chem Kinet. 1993;25(4):239–247.
- Cheng T, Li X, Li Y, Liu Z, Wang R. Comparative assessment of scoring functions on a diverse test set. J Chem Inf Model. 2009;49(4):1079–1093.
- Sándor M, Kiss R, Keseru GM. Virtual fragment docking by glide: a validation study on 190 protein − fragment complexes. J Chem Inf Model. 2010;50(6):1165–1172.
- Mammoli A, Bianconi E, Ruta L, Riccio A, Bigiotti C, Souma M, Carotti A, Rossini S, Suvieri C, Pallotta MT, et al. Critical Assessment of a structure-based screening campaign for IDO1 inhibitors: tips and pitfalls. Int J Mol Sci. 2022;23(7):3981.
- de Araujo IG, Pattaro-Júnior JR, Barbosa CG, Philippsen GS, Silva AR, Ioshino RS, Moraes CB, Freitas-Junior LH, Barros L, Peralta RM, et al. Potential of plant extracts in targeting SARS-CoV-2 main protease: an in vitro and in silico study. J Biomol Struct Dyn. 2023:1–10.
- Pattaro-Júnior JR, Araújo IG, Moraes CB, Barbosa CG, Philippsen GS, Freitas-Junior LH, Guidi AC, de Mello JCP, Peralta RM, Fernandez MA, et al. Antiviral activity of Cenostigma pluviosum var. peltophoroides extract and fractions against SARS-CoV-2. J Biomol Struct Dyn. 2023;41(15):7297–7308.
- Völgyi G, Ruiz R, Box K, Comer J, Bosch E, Takács-Novák K. Potentiometric and spectrophotometric pKa determination of water-insoluble compounds: validation study in a new cosolvent system. Anal Chim Acta. 2007;583(2):418–428.
- Garidel P, Hegyi M, Bassarab S, Weichel M. A rapid, sensitive and economical assessment of monoclonal antibody conformational stability by intrinsic tryptophan fluorescence spectroscopy. Biotechnol J. 2008;3(9–10):1201–1211.
- Duhr S, Braun D. Why molecules move along a temperature gradient. Proc Natl Acad Sci USA. 2006;103(52):19678–19682.
- Abad-Zapatero C. Ligand efficiency indices for effective drug discovery. Expert Opin Drug Discov. 2007;2(4):469–488.
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49(D1):D1388–D1395.
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47(7):1739–1749.
- Gidari A, Sabbatini S, Bastianelli S, Pierucci S, Busti C, Bartolini D, Stabile AM, Monari C, Galli F, Rende M, et al. SARS-CoV-2 survival on surfaces and the effect of UV-C light. Viruses. 2021;13(3):408.
- Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol. 1938;27(3):493–497.
- Gidari A, Sabbatini S, Bastianelli S, Pierucci S, Busti C, Monari C, Luciani Pasqua B, Dragoni F, Schiaroli E, Zazzi M, et al. Cross-neutralization of SARS-CoV-2 B.1.1.7 and P.1 variants in vaccinated, convalescent and P.1 infected. J Infect. 2021;83(4):467–472.
- Lai A, Bergna A, Caucci S, Clementi N, Vicenti I, Dragoni F, Cattelan AM, Menzo S, Pan A, Callegaro A, et al. Molecular tracing of SARS-CoV-2 in Italy in the first three months of the epidemic. Viruses. 2020;12(8):798.
- Abdelnabi R, Foo CS, Jochmans D, Vangeel L, De Jonghe S, Augustijns P, Mols R, Weynand B, Wattanakul T, Hoglund RM, et al. The oral protease inhibitor (PF-07321332) protects Syrian hamsters against infection with SARS-CoV-2 variants of concern. Nat Commun. 2022;13(1):719.
- Gidari A, Sabbatini S, Pallotto C, Bastianelli S, Pierucci S, Busti C, Schiaroli E, Francisci D. Nelfinavir: an old ally in the COVID-19 fight? Microorganisms. 2022;10(12):2471.