Abstract
Objective
To demonstrate the prevalence of maternal mosaic monosomy X (MMXO) in a cohort of pregnant women in Vietnam.
Methods
All 105,594 singleton pregnant women undergoing noninvasive prenatal screening (NIPS) between January 2019 and February 2021 in Vietnam were analyzed by measuring discordance between size- and count-based z-scores for chromosome X (ChrX) to identify suspected cases of MMXO and validated by fluorescence in situ hybridization (FISH) on maternal blood.
Results
We identified 295 (0.279%) suspected MMXO cases. After FISH analysis, MMXO was confirmed in 125 cases (42.37%), revealing the MMXO prevalence of 0.118% (95% CI: 0.097–0.139%) in this cohort.
Conclusion
We found a relatively high prevalence of MMXO in Vietnamese pregnant women and demonstrated a strong influence of MMXO on the ChrX z-score using a count-based method, resulting in false positives. The size-based method is not sensitive to MMXO and therefore achieves higher PPV.
Noninvasive prenatal screening (NIPS) uses cell-free fetal DNA (cffDNA) in maternal plasma to screen for aneuploidies. It detects fetal aneuploidies with high clinical specificity, outperforming conventional screening, and is recommended by the American College of Medical Genetics and Genomics for common aneuploidies [Citation1]. NIPS of sex chromosome aneuploidy is also recommended, with pretest counseling about false positives. Fetal sex chromosome aneuploidy was found in 1.1% of 18,161 cases, with monosomy X (MX) being the most prevalent (148 cases, 0.81%) [Citation2]. However, of the 44 MX cases with clinical follow-up, there were 35 false-positives, yielding a false-positive rate (FPR) of 0.65% and a positive predictive value (PPV) of 20.5% [Citation2]. Possible causes for high FPR include confined placental mosaicism, a co-twin with MX demise, maternal constitutional MX (full or mosaic), and maternal somatic mosaic MX due to age-related X chromosome loss. A SNP-based NIPS approach detected maternal X chromosome abnormalities with a PPV of 94.3%, of which 54.7% were mosaicism for MX [Citation3]. We previously reported a method identifying maternal mosaic MX (MMXO) by measuring discordance between size- and count-based z-scores for chromosome X (ChrX) [Citation4]. This study retrospectively identified MMXO cases to estimate MMXO prevalence in Vietnamese pregnant women.
Women with a singleton pregnancy (N = 105,594) underwent NIPS between January 2019 and February 2021 at Gene Solutions—Medical Genetics Institute. All participants received pretest counseling and provided informed consent for their data to be used for research. Peripheral blood (10 ml) was collected, and cell‐free DNA was extracted (MagMAX Cell‐Free DNA Isolation Kit; Thermo Fisher Scientific). Libraries were prepared (NEBNext Ultra II DNA Library Prep Kit; New England Biolabs) and sequenced (NextSeq platform using 2 × 75‐bp Kit; Illumina). All sequencing data were analyzed by the triSure method as previously described [Citation4,Citation5]. TriSure is a size-based method that calculates the difference in proportion (DP) of fetal derived (≤150 bp) over maternal-derived (≥170 bp) fragments between the target chromosome and all other chromosomes [Citation5]. The DP of each chromosome is then used to determine chromosome specific z-score by comparing it with the DP from a reference set. A fetal monosomy X case was defined as having a ChrX z-score calculated by the triSure (ztriSure) method [Citation5] below −3. A suspected MMXO case was defined as having a ChrX z-score calculated by a count-based method (NIPTeR package [Citation6], zNIPTeR) below −3.0 while the corresponding z-scores by the triSure (ztriSure) method [Citation5] were above −3.0.
Suspected MMXO cases (295; 0.279%) were identified by the combined method (zNIPTeR < −3 and ztriSure > −3) and validated by fluorescence in situ hybridization (FISH) (Supplemental Table 1). MMXO was confirmed in 125 (42.37%), and 20 were other types of maternal sex chromosome mosaicism, including XX/XXX, XX/partial Y, and XX/XY. FISH results did not detect any chromosomal abnormalities in the remaining 150 cases. Because FISH were performed on maternal blood, we could not rule out the possibility that these cases were maternal somatic mosaic MX. Importantly, all infants were born healthy, consistent with the ChrX ztriSure > −3.0.
We identified 242 cases (0.229%) as at high-risk of fetal MX (fMX) (zNIPTeR < −3 and ztriSure < −3). Thirty-three underwent amniocentesis or chronic villus sampling and FISH analysis, revealing 14 cases and 19 false positives (PPV = 42.4%, 95%CI 25.5–59.3%). Using this PPV, we estimated that the number of fMX cases was 103 (0.098%; 95%CI 62–144).
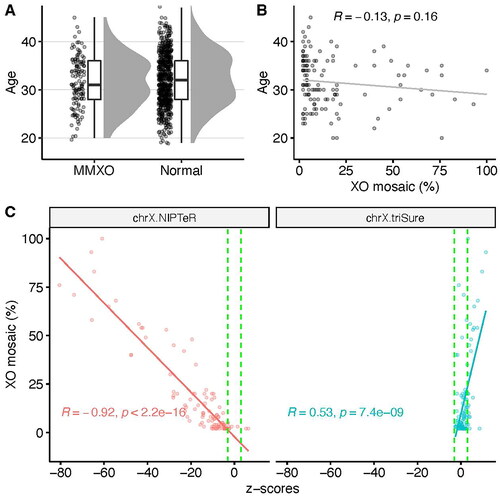
MMXO is a major biological factor contributing to NIPS’ high false positive rates of fMX, but few studies report its prevalence. Using a combination of count- and size-based methods, we identified 295 cases (0.279%) suspected of having MMXO and validated with maternal blood FISH analysis, revealing MMXO prevalence as 0.118% (95% CI: 0.097–0.139%). This is higher than previously reported in a study that found 40 cases of MMXO in 141,916 women (0.03%) [Citation7], but lower than in another study of 906 women, three of which were MMXO (0.3%) [Citation8]. MMXO can be constitutional or age-related somatic X chromosome loss. Because our FISH analysis was performed on blood cells, maternal somatic mosaic MX was unlikely to be identified. Indeed, our data show that the maternal age distribution of confirmed MMXO cases was like normal cases (Kolmogorov–Smirnov test, p = .497, ) with no significant difference between the two groups (Mann–Whitney test, p = .3689). Furthermore, no correlation was found between age and percentage of XO mosaicism (Pearson’s R = −0.13, p = .16, ). This suggested that the prevalence of confirmed MMXO reported here is probably underestimated, and there were likely undetected age-related somatic MMXO in the 150 cases suspected of MMXO not confirmed by FISH.
Figure 1. (A) Maternal age distribution of FISH-confirmed MMXO cases compared to normal cases. For visualization purposes, only 500 randomly selected normal cases were plotted. (B) Low correlation between maternal age and percent of maternal XO mosaicism. (C) Strong negative correlation between percent of maternal XO mosaicism and z-scores of ChrX calculated by count-based method (left panel), demonstrating the influence of maternal XO mosaicism, while the triSure method (right panel) is not sensitive to the confounding effect of maternal XO mosaicism.
Our data suggested that a count-based method might report approximately 537 (=295 + 242, zNIPTeR < −3) cases as fMX (0.508%), of which 434 were false positive with a FPR of 0.41%. This FPR is realistic and slightly lower than previously reported (0.65%) [Citation2]. Encouragingly, the triSure method only counted 242 cases as fXO (295 cases of suspected MMXO showed ztriSure > −3 and were not considered fXO by triSure alone), reducing the false positive rate to 0.13% (assuming PPV = 42.4% in fXO by triSure). A recent study on 94,085 women with a singleton pregnancy reported 49 true positives and 141 false positives (0.15% FPR, PPV = 25.8%) [Citation9], while another study of 25,517 pregnant women reported an FPR of 0.09% and PPV of 12% [Citation10], indicating that the triSure method achieves similar FPR but higher PPV. Previously, Wang et al. found that 8.06% of NIPS MX cases were MMXO by applying massively parallel sequencing to measure the degree of chromosome mosaicism from maternal white blood cells [Citation11]. However, this approach added the extra cost of testing all NIPS MX cases. In contrast, the triSure method is not sensitive to the influence of MMXO and therefore reduces the number of cases requiring invasive procedures or maternal blood cell karyotype analysis.
To better demonstrate the influence of MMXO on z-scores of ChrX, we used the FISH-confirmed MMXO samples to plot the percentage of XO mosaicism against ChrX z-scores calculated by either NIPTeR or the triSure method (). There was a strong negative correlation between the percentage of mosaicism and z-scores by NIPTeR (Pearson’s R = −0.92, p < 2.2 × 10−16), suggesting that the higher the percent of ChrX loss, the stronger its influence on z-scores by NIPTeR. In contrast, the z-scores by triSure were all above −3.0, indicating that none of these samples were identified.
In conclusion, our study highlights a relatively high prevalence of MMXO in pregnant women in Vietnam. We also demonstrated the influence of MMXO on the ChrX z-score for using a count-based method, resulting in false positives. Size-based method, in contrast, is not sensitive to the confounding effect of MMXO and therefore achieves higher PPV.
Ethical approval
The studies involving human participants were reviewed and approved by the institutional ethics committee of the University of Medicine and Pharmacy, Ho Chi Minh City, Vietnam (approval number 164/HDDD). Written informed consent to participate in this study was provided by the participants.
Supplemental Material
Download MS Word (14.6 KB)Acknowledgments
We would like to acknowledge the contribution of all pregnant women who agreed to participate in this study. We thank Angela Marie Jansen, Ph.D., MHS of Angela Jansen and Associates, for assistance in editing the manuscript.
Disclosure statement
TTTN, NTKN, CTN, TDN, HG, HST, and MDP are employed by Gene Solutions, a company providing NIPS using the triSure method. No conflict of interest was reported by the remaining authors.
Data availability statement
All data are presented in this manuscript.
Additional information
Funding
References
- Gregg AR, Skotko BG, Benkendorf JL, et al. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genet Med. 2016;18(10):1056–1065.
- Bianchi DW, Parsa S, Bhatt S, et al. Fetal sex chromosome testing by maternal plasma DNA sequencing: clinical laboratory experience and biology. Obstet Gynecol. 2015;125(2):375–382.
- Martin KA, Samango-Sprouse CA, Kantor V, et al. Detection of maternal X chromosome abnormalities using single nucleotide polymorphism–based noninvasive prenatal testing. Am J Obstet Gynecol MFM. 2020;2(3):100152.
- Phan M, Vo BT, Nguyen TV, et al. Reducing false positive rate of fetal monosomy X in non‐invasive prenatal testing using a combined algorithm to detect maternal mosaic monosomy X. Prenat Diagn. 2019;39(4):324–327.
- Phan M-D, Nguyen TV, Trinh HNT, et al. Establishing and validating noninvasive prenatal testing procedure for fetal aneuploidies in Vietnam. J Matern Fetal Neonatal Med. 2019;32(23):4009–4015.
- Johansson LF, de Boer EN, de Weerd HA, et al. Novel algorithms for improved sensitivity in non-invasive prenatal testing. Sci Rep. 2017;7(1):1838.
- Samango-Sprouse C, Kırkızlar E, Hall MP, et al. Incidence of X and Y chromosomal aneuploidy in a large child bearing population. PLOS One. 2016;11(8):e0161045.
- Shubina J, Trofimov DY, Barkov IY, et al. In silico size selection is effective in reducing false positive NIPS cases of monosomy X that are due to maternal mosaic monosomy X. Prenat Diagn. 2017;37(13):1305–1310.
- Liang D, Cram DS, Tan H, et al. Clinical utility of noninvasive prenatal screening for expanded chromosome disease syndromes. Genet Med. 2019;21(9):1998–2006.
- Pang Y, Wang C, Tang J, et al. Clinical application of noninvasive prenatal testing in the detection of fetal chromosomal diseasxes. Mol Cytogenet. 2021;14(1):31.
- Wang Y, Chen Y, Tian F, et al. Maternal mosaicism is a significant contributor to discordant sex chromosomal aneuploidies associated with noninvasive prenatal testing. Clin Chem. 2014;60(1):251–259.

