Abstract
Objectives
To explore the prenatal clinical utility of chromosome microarray analysis (CMA) for polyhydramnios and evaluate the short and long-term prognosis of fetuses with polyhydramnios.
Methods
A total of 600 singleton pregnancies with persistent polyhydramnios from 2014 to 2020 were retrospectively enrolled in this study. All cases received amniocentesis and were subjected to CMA results. All cases were categorized into two groups: isolated polyhydramnios and non-isolated polyhydramnios [with soft marker(s) or with sonographic structural anomalies]. All fetuses were followed up from 6 months to five years after amniocentesis to acquire short and long-term prognosis.
Results
The detection rates of either aneuploidy or pathogenic copy number variants in fetuses with non-isolated polyhydramnios were significantly higher than those with isolated polyhydramnios (5.0 vs. 1.5%, p = 0.0243; 3.6 vs. 0.8%, p = 0.0288). The detection rate of total chromosomal abnormalities in the structural abnormality group was significantly higher than that in the isolated group (10.0 vs. 2.3%, p = 0.0003). In the CMA-negative cases, the incidence of termination of pregnancy, neonatal and childhood death, and non-neurodevelopmental disorders in fetuses combined with structural anomalies was significantly higher than that in fetuses with isolated polyhydramnios (p < 0.05). We did not observe any difference in the prognosis between the isolated group and the combined group of ultrasound soft markers. In addition, the risk of postnatal neurodevelopmental disorders was also consistent among the three groups (1.6 vs. 1.3 vs. 1.8%).
Conclusion
For low-risk pregnancies, invasive prenatal diagnosis of isolated polyhydramnios might be unnecessary. CMA should be considered for fetuses with structural anomalies. In CMA-negative cases, the prognosis of fetuses with isolated polyhydramnios was good, and polyhydramnios itself did not increase the risk of postnatal neurological development disorders. The worse prognosis mainly depends on the combination of polyhydramnios with structural abnormalities.
Introduction
Polyhydramnios is an abnormal increase in amniotic fluid volume that is typically diagnosed in the second or third trimester, and it affects ∼1–2% of all pregnancies [Citation1]. Physiological impairment of fetal swallowing or overproduction of fetal urine due to a high-output cardiac state, renal abnormality, or osmotic fetal diuresis can result in polyhydramnios. The five common pathological causes of polyhydramnios are maternal diabetes mellitus, fetal anomalies, congenital infection, placental tumors, and iso-immunization [Citation2]. No specific etiology can be identified in ∼50–60% of cases [Citation1].
The identification of polyhydramnios should prompt the elucidation of the underlying etiology. Previous reports examining the association between polyhydramnios and genetic anomalies have shown that polyhydramnios might be associated with an increased risk of abnormal karyotypes [Citation3–5]. Chromosomal microarray analysis (CMA) detects an imbalance in DNA copy number, which is referred to as a copy number variant (CNV) [Citation6]. In cases with structural anomalies found during prenatal imaging examinations, CMA can detect an additional 5.6% of pathogenic CNVs in isolated defects and 9.1% of pathogenic CNVs in multiple defects [Citation7]. To date, the role of prenatal copy number variation testing in cases with polyhydramnios has been studied in a limited number of literatures [Citation8–10]. However, the findings are not entirely consistent. Especially in isolated polyhydramnios cases, the detection rate of pathogenic or likely pathogenic CNVs varied from 2.5 to 5.7%. In addition, several studies summarized the pregnancy outcomes of women with polyhydramnios and suggested there was an increased risk of adverse outcomes in pregnancies complicated by idiopathic polyhydramnios [Citation11–13]. A recent meta-analysis concluded that idiopathic polyhydramnios significantly increased the risk of intrauterine fetal demise and neonatal death [Citation11]. However, fetal outcomes after prenatal diagnosis with CMA, especially long-term prognosis, are still unclear.
Therefore, in this retrospective study, we systematically investigated chromosomal aberrations in singleton pregnancies with isolated and non-isolated polyhydramnios, either with soft markers or structural anomalies. Furthermore, short and long-term prognosis were evaluated comprehensively based on the follow-up information of the fetuses receiving CMA tests.
Methods
Study population
A total of 600 singleton pregnancies with persistent polyhydramnios detected by ultrasound at West China Second University Hospital, Sichuan University, from September 2014 to December 2020 were included in this retrospective study. Pretest counseling was provided by trained clinical geneticists. All fetal samples were obtained by amniocentesis at gestational ages ranging from 20 to 36 weeks. Maternal age was between 18 and 45 years. All women <35 years of age had received common aneuploidy screening, either through serum screening or noninvasive prenatal screening, during routine obstetric examination before amniocentesis, and the results categorized all pregnancies as low risk. Written informed consent was obtained from all pregnant women before testing. This study was approved by the Medical Ethics Committee of West China Second University Hospital, Sichuan University, and the research was conducted in accordance with relevant guidelines and clinical norms.
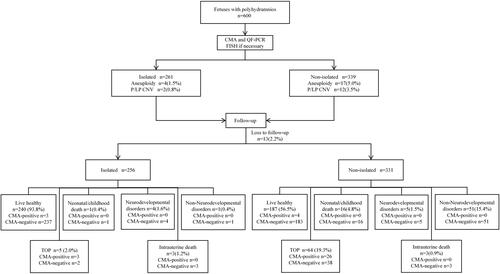
All cases were categorized into two groups: isolated polyhydramnios (n = 261) and non-isolated polyhydramnios (n = 339) [with soft marker(s) (n = 88) or with sonographic structural anomalies (n = 251)]. The study procedure is shown in .
Definition and degree of polyhydramnios
Amniotic fluid was assessed by ultrasonography using either of the two semi-quantitative methods: (1) single deepest vertical pocket (DVP) of amniotic fluid, with polyhydramnios defined as ≥8 cm or (2) amniotic fluid index (AFI), with polyhydramnios defined as ≥25 cm. The degree of polyhydramnios is frequently categorized as mild, moderate, or severe based on an AFI of 25.0–29.9, 30.0–34.9, and ≥35 cm, respectively, or a DVP of 8–11.9, 12–15.9, or ≥16 cm, respectively [Citation8,Citation14,Citation15].
CMA, quantitative fluorescent polymerase chain reaction (QF-PCR), and fluorescence in situ hybridization (FISH)
The experimental procedures were performed as described in our previous study [Citation16]. The initial 2 ml of amniotic fluid (AF) was removed to prevent maternal cell contamination (MCC). In case of suspected MCC, DNA was extracted from the cultured AF. If MCC was excluded, DNA was extracted immediately from the uncultured AF using a QIAamp® DNA Blood Mini Kit (QIAGEN GmbH, Hilden, Germany). All samples were subjected to QF-PCR detection to identify MCC and to reconfirm common autosome chromosome aneuploidy using a 21, 18, and 13 trisomy and sex chromosome polyploidy detection kit (fluorescence PCR-capillary electrophoresis) (DAAN GENE, Guangzhou, China), following the manufacturer’s protocol. PCR fragments were separated by capillary electrophoresis (3500 Genetic Analyzer, Life Technologies, CA, USA), and the data were analyzed using GeneMapper® (version 4.1, Applied Biosystems, Waltham, MA, USA).
All samples were screened using a CytoScan 750K array (Affymetrix Inc., Santa Clara, CA, USA). The sensitivity and specificity have been universally acknowledged, and CNVs ≥100 kb across the genome could be reliably detected. The experimental procedures were performed according to the manufacturer’s standard protocol (Affymetrix Inc., Santa Clara, CA, USA). The results were analyzed by two clinical geneticists using Chromosome Analysis Suite (ChAS) software (Affymetrix, Inc., Santa Clara, CA, USA). Sex chromosome aneuploidy was confirmed by FISH. The results were determined using in-house databases and publicly available CNV databases, including the Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER; http://decipher.sanger.ac.uk), GeneReviews®, Database of Genomic Variants (DGV; http://projects.tcag.ca/variation), Online Mendelian Inheritance in Man (OMIM; http://www.omim.org), and ClinGen (https://www.clinicalgenome.org/). The databases are occasionally not updated with the latest literature; consequently, PubMed was also included in the data analysis process. In our study, conventional cytogenetic analysis was not performed in addition to CMA. We categorized CNVs as benign, pathogenic, or variants of uncertain significance (VOUS) according to the American College of Medical Genetics (ACMG) standards and guidelines for the interpretation and reporting of postnatal constitutional CNVs [Citation17]. If VOUS were detected in the fetal sample, peripheral blood was collected from both parents and the results were further analyzed to regrade the CNVs. Clinical geneticists at our prenatal diagnosis center offered counseling to the participants based on the array results.
Postnatal follow-up assessments
Clinical follow-up assessments via telephone or medical records were scheduled and performed from 6 months to five years after amniocentesis to evaluate the pregnancy outcomes and circumstances after birth, including postpartum image findings and developmental details.
Statistical analysis
GraphPad Prism, version 9 (GraphPad Software Inc., CA, USA), was used for the statistical analysis. We used the chi-square test or Fisher’s exact test to identify significant differences in the expected frequencies between the two groups. A value of p < 0.05 was considered statistically significant in two-sided tests.
Results
Prevalence of chromosomal aberrations in fetuses with polyhydramnios
The overall prevalence of chromosomal aberrations in fetuses with polyhydramnios was 5.8% (35/600). Among all cases, 21 (3.5%) fetuses showed common aneuploidies involving chromosome 18, 21, and sex chromosomes, while 14 (2.3%) fetuses were identified based on pathogenic copy number variants (pCNVs). Four types of microdeletion/microduplication syndromes were identified in 10 samples, including 22q11.2 deletion syndrome, renal cysts and diabetes syndrome (RCAD), hereditary neuropathy with liability to pressure palsies (HNPP), and Miller-Dieker syndrome. The study procedure is shown in .
Among the 261 pregnancies with isolated polyhydramnios, chromosomal aberrations were detected in six cases (2.3%), four fetuses (1.5%) showed common aneuploidies (two trisomy X and two 47,XYY), and two fetuses (0.8%) showed pCNVs [22q11.2 deletion syndrome and Hereditary neuropathy with liability to pressure palsies (HNPP)] ( and Supplementary Table 1). Among the 339 pregnancies with non-isolated polyhydramnios, total chromosomal aberrations, common aneuploidies, and pCNVs were detected in 29 (8.6%), 17 (5.0%), and 12 cases (3.5%), respectively (). The incidence of common aneuploidies and pCNVs in isolated polyhydramnios was significantly lower than that in non-isolated polyhydramnios (1.5 vs. 5.0%, p = 0.0243; 0.8 vs. 3.5%, p = 0.0288, respectively). Moreover, we found no pathogenic CNV result in cases with soft markers (Supplementary Table 2).
Table 1. Comparison of detection rates of chromosomal anomalies between isolated and non-isolated polyhydramnios.
In 251 cases with structural malformations, chromosomal aberrations, aneuploidies, and pCNVs were detected in 25 (10.0%), 13 (5.2%), and 12 (4.8%) cases, respectively (). Among the fetuses with pCNVs, all three RCAD cases were presented with hyperechogenic kidneys, and both 22q11.2 microdeletion syndrome cases were found with congenital heart disease. Detailed information on clinically significant CMA findings in cases combined with structural malformations was shown in Supplementary Table 3.
The detection rates of total chromosomal aberrations in fetuses with polyhydramnios combined with sonographic structural anomalies were significantly higher than those of isolated polyhydramnios while those with soft markers were not (10.0 vs. 2.3%, p = 0.0003; 4.5 vs. 2.3%, p = 0.2790) ().
Subgroup analysis of different severities of polyhydramnios
In the 600 fetuses with polyhydramnios, the percentages of mild, moderate, and severe polyhydramnios were 89.0% (534/600), 9.3% (56/600), and 1.7% (10/600), respectively. Moreover, in fetuses with mild, moderate, and severe polyhydramnios, the incidence of common aneuploidies and pCNVs was 5.4% (29/534), 8.9% (5/56), and 10% (1/10), respectively. However, no statistical differences were observed between any two groups (Supplementary Table 4).
Clinical follow-up assessment
The pregnancy outcomes of all fetuses with polyhydramnios were followed-up over a period of 6 months to five years. A total of 13 patients (2.2%) were lost to follow-up; thus, the pregnancy outcomes were evaluated in 587 fetuses (587/600, 97.8%).
Except for one fetus confirmed with trisomy X and three fetuses confirmed with 47,XYY syndrome, the other 17 fetuses with common aneuploidies were subjected to termination of pregnancy (TOP). Except for one fetus confirmed with RCAD and one fetus confirmed with HNPP, the other 11 fetuses with pCNVs were subjected to TOP. Among the five fetuses with chromosomal anomalies that survived till birth, the parents indicated that the children’s conditions were all satisfactory based on telephone follow-up. The detailed information on the prognosis of these CMA-positive fetuses is shown in Supplementary Tables 1–3.
Table 2. Comparison of pregnancy outcomes between fetuses with isolated and non-isolated polyhydramnios with CMA negative results.
Table 3. Poor prognostic outcomes in fetuses with normal CMA results in isolated group.
The rate of CMA-negative fetuses was 92.8% (557/600), including 253 isolated polyhydramnios and 304 non-isolated polyhydramnios, and 2.3% (13/557) were lost to follow-up. In these CMA-negative cases, the incidence of TOP, neonatal and childhood death, and non-neurodevelopmental disorders in fetuses combined with structural anomalies were significantly higher than that in fetuses with isolated polyhydramnios (p < 0.05). On the other hand, we did not observe any difference in the prognosis between the isolated group and the combined group of ultrasound soft markers. In addition, the risk of postnatal neurodevelopmental disorders was also consistent among the three groups. The detailed information on the prognosis of those CMA-negative fetuses is shown in .
In fetuses with negative prenatal CMA results in the isolated group, new diseases were found in 6 fetuses after birth, and one of them died of CHARGE syndrome. The total anomalous pulmonary venous connection was found in one neonate by echocardiography. The surgical outcome was good and the patient was healthy during follow-up. Four children had developmental abnormalities of the nervous system. See for details.
Discussion
Principal findings
Our study included a large series of cases to establish the correlations of common aneuploidies and pCNVs with polyhydramnios, and pregnancy outcomes were obtained from 587 cases from 6 months to 5 years. The detection rate of chromosome anomalies of fetuses with polyhydramnios in the non-isolated group was significantly higher than that in the isolated group. Among the CMA-negative fetuses, the pregnancy outcomes of fetuses with isolated polyhydramnios and non-isolated polyhydramnios were significantly different except for the condition of intrauterine death and neurodevelopmental disorders.
Clinical implications
Whether isolated polyhydramnios increases the risk of chromosomal abnormalities is one of our focuses. Sagi-Dain et al. [Citation8] summarized the characteristics of 742 fetuses with polyhadramnios and found that the chromosomal abnormality detection rate for isolated polyhydramnios was 3.1% (19/623, including 17 pCNVs and two chromosomal aneuploidies), which was significantly higher than that in the control group of fetuses with normal ultrasound (1.4%, 78/5541). However, the degree of polyhydramnios and maternal age were not known in some patients. Our findings are different from this conclusion. In our cohort, the chromosomal abnormality detection rate in the isolated group was similar to that of normal ultrasound (2.3%, 6/261, two cases with pCNVs, and four cases with chromosomal aneuploidies). The four cases of chromosomal aneuploidies included two trisomy X and two 47,XYY, not the common autosomal aneuploidies. These two sex chromosomal aneuploidies showed relatively normal cognitive and behavioral phenotypes [Citation18,Citation19], and most pregnant women with fetuses presenting these conditions choose to continue their pregnancy. There were a few other studies exploring the association of chromosome anomalies and isolated polyhydramnios, and the detection rate varied from 2.6 to 5% [Citation9,Citation10]. However, neither study indicated whether the participants were screened for common chromosomal aneuploidies before amniocentesis and what the results were. In our study, cases with a high risk of noninvasive prenatal screening or serologic screening have been excluded, and all cases younger than 35 years old were low-risk populations on common autosomal aneuploidy screening tests. Therefore, for low-risk pregnant women, we suggest isolated polyhydramnios do not increase the risk of chromosome abnormalities.
Sagi-Dain et al. [Citation8] found that the pCNV detection rate for non-isolated polyhydramnios was 6.7% (8/119), which was slightly higher than the rate in our study (3.6%, 12/336). In the former study, the exact meaning of non-isolated polyhydramnios was not clearly described in the text; however, in our study, both combined ultrasound soft markers and structural abnormalities were grouped into non-isolated polyhydramnios. The detection rates of aneuploidy and pCNVs in polyhydramnios with soft markers and structural anomalies (4.5 and 0%; 5.2 and 4.8%, respectively) were both comparable to those including our previous studies focused on the use of CMA for soft markers and structural anomalies [Citation7,Citation20–22].
In the study of Sagi-Dain et al. [Citation8], more than one case of 17q12 deletion and 22q11.2 deletion syndrome were found in the isolated polyhydramnios group. This was quite different from our results which showed that all cases with these deletion syndromes were found to have additional sonographic findings. In our study, three cases of 17q12 deletion were identified by CMA, and all of these were diagnosed as polyhydramnios combined with a hyperechogenic kidney. Jing et al. [Citation23] found 12 cases with 17q12 deletion from ∼6000 invasive cytogenetic tests. In their retrospective study, variable kidney abnormalities were found by ultrasound in all of the 17q12 deletion cases, with bilateral or unilateral hyperechogenic kidneys being the most common findings. In another study, 12 in 3320 fetuses were detected to have 17q12 deletion, all characterized as renal abnormalities in which 11 cases with bilateral hyperechogenic kidneys and one case with renal cyst [Citation24]. Therefore, we suggest that when polyhydramnios are found during pregnancy, a detailed and comprehensive ultrasound examination is necessary.
Although reports of idiopathic polyhydramnios associated with perinatal mortality have been inconsistent, most results suggest that idiopathic polyhydramnios is associated with specific adverse outcomes, such as a higher rate of cesarean delivery, macrosomia, and NICU admissions [Citation1,Citation12,Citation25–27]. However, these studies focused on delivery outcomes, and no studies have performed long-term follow-up of fetuses with polyhydramnios after birth. We followed up fetuses with polyhydramnios for 6 months to 5 years after birth and obtained more detailed follow-up data. We focused on the postnatal development of the nervous system. Our follow-up data suggest that the proportion of neurodevelopmental disorders in the isolated group was basically the same as that in the non-isolated group, showing no significant difference. These results (1.3–1.8%) were comparable to the reported global prevalence of intellectual disability in children (1983 cases per 100,000 children) [Citation28]. Therefore, we conclude that the risk of postnatal neurodevelopmental disorders in fetuses experiencing polyhydramnios during pregnancy might not be significantly higher than that in the general population. The pregnancy outcome of other fetuses with negative CMA results was mainly dependent on the presence and severity of concomitant structural abnormalities. In the isolated group, we found two non-isolated cases postnatally. One fetus was found to have multiple malformations after birth and was diagnosed with CHARGE syndrome by whole exome sequencing, and died in the neonatal period. The other fetus was diagnosed with total anomalous pulmonary venous connection by echocardiography due to shortness of breath and cyanosis after birth. He underwent cardiac surgery and was in good health at follow-up. The above follow-up results showed that the prognosis of isolated polyhydramnios is good if no other structural abnormalities were present. However, prenatal determination of whether it is truly isolated polyhydramnios is challenging due to the limitations of ultrasound technology.
Strengths and limitations
The major strength of this study is the large size of the cohort from a single center and the further stratification of non-isolated polyhydramnios into combined ultrasound soft markers and structural abnormalities. In addition, long-term follow-up results can provide updated information for clinical genetic counseling for polyhydramnios. This provides more details for prenatal genetic counseling. We acknowledge the limitations of this review. This was a retrospective study, the follow-ups were mainly performed over the telephone, and some patients were lost to follow-up, which might have introduced bias in the study results.
Conclusion
The detection rates of chromosomal abnormalities in fetuses with polyhydramnios combined with structural anomalies were significantly higher than those in isolated polyhydramnios. When common chromosomal trisomy screening during pregnancy is low risk, an invasive prenatal diagnosis for isolated polyhydramnios might not significantly increase the detection rate of chromosome anomalies. CMA should be performed when polyhydramnios are combined with structural anomalies. Our results also indicated that the prognosis of fetuses with isolated polyhydramnios was good, and polyhydramnios itself did not increase the risk of postnatal neurological development disorders.
Supplemental Material
Download Zip (62.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data is available on request from the authors.
Additional information
Funding
References
- Magann EF, Chauhan SP, Doherty DA, et al. A review of idiopathic hydramnios and pregnancy outcomes. Obstet Gynecol Surv. 2007;62(12):1–9. doi: 10.1097/01.ogx.0000290349.58707.e0.
- Dashe JS, Pressman EK, Hibbard JU, et al. SMFM consult series #46: evaluation and management of polyhydramnios. Am J Obstet Gynecol. 2018;219(4):B2–B8. doi: 10.1016/j.ajog.2018.07.016.
- Shimada S, Yamada H, Hoshi N, et al. Specific ultrasound findings associated with fetal chromosome abnormalities. Congenit Anom. 2009;49(2):61–65. doi: 10.1111/j.1741-4520.2009.00224.x.
- Enzensberger C, Pulvermacher C, Degenhardt J, et al. Fetal loss rate and associated risk factors after amniocentesis, chorionic villus sampling and fetal blood sampling. Ultraschall Med. 2012;33(7):E75–E79. doi: 10.1055/s-0031-1299388.
- Sagi-Dain L, Sagi S. Chromosomal aberrations in idiopathic polyhydramnios: a systematic review and meta-analysis. Eur J Med Genet. 2015;58(8):409–415. doi: 10.1016/j.ejmg.2015.06.010.
- Levy B, Wapner R. Prenatal diagnosis by chromosomal microarray analysis. Fertil Steril. 2018;109(2):201–212. doi: 10.1016/j.fertnstert.2018.01.005.
- Wapner RJ, Martin CL, Levy B, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012;367(23):2175–2184. doi: 10.1056/NEJMoa1203382.
- Sagi-Dain L, Singer A, Falik-Zaccai T, et al. The effect of polyhydramnios degree on chromosomal microarray results: a retrospective cohort analysis of 742 singleton pregnancies. Arch Gynecol Obstet. 2021;304(3):649–656. doi: 10.1007/s00404-021-05995-y.
- Shi P, Hou Y, Chen D, et al. Estimate of genetic variants using CNV-seq for fetuses with oligohydramnios or polyhydramnios. Mol Genet Genomic Med. 2023;11(1):e2089.
- Wu X, Li Y, Lin N, et al. Evaluation of genetic variants using chromosomal microarray analysis for fetuses with polyhydramnios. BMC Med Genomics. 2022;15(1):73. doi: 10.1186/s12920-022-01224-w.
- Pagan M, Magann EF, Rabie N, et al. Idiopathic polyhydramnios and pregnancy outcome: systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2023;61(3):302–309. doi: 10.1002/uog.24973.
- Luo QQ, Zou L, Gao H, et al. Idiopathic polyhydramnios at term and pregnancy outcomes: a multicenter observational study. J Matern Fetal Neonatal Med. 2017;30(14):1755–1759. doi: 10.1080/14767058.2016.1224835.
- Wax JR, Cartin A, Craig WY, et al. Transient idiopathic polyhydramnios: maternal and perinatal outcomes: maternal and perinatal outcomes. J Ultrasound Med. 2022;41(11):2859–2866. doi: 10.1002/jum.15974.
- Dashe JS, McIntire DD, Ramus RM, et al. Hydramnios: anomaly prevalence and sonographic detection. Obstet Gynecol. 2002;100(1):134–139. doi: 10.1097/00006250-200207000-00021.
- Kouamé N, N’goan-Domoua AM, Nikiéma Z, et al. Polyhydramnios: a warning sign in the prenatal ultrasound diagnosis of foetal malformation? Diagn Interv Imaging. 2013;94(4):433–437. doi: 10.1016/j.diii.2013.01.002.
- Zhang Z, Hu T, Wang J, et al. Prenatal diagnostic value of chromosomal microarray in fetuses with nuchal translucency greater than 2.5 mm. Biomed Res Int. 2019;2019:6504159. doi: 10.1155/2019/6504159.
- Riggs ER, Andersen EF, Cherry AM, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the clinical genome resource (ClinGen). Genet Med. 2020;22(2):245–257. doi: 10.1038/s41436-019-0686-8.
- Tartaglia NR, Howell S, Sutherland A, et al. A review of trisomy X (47,XXX). Orphanet J Rare Dis. 2010;5(1):8. doi: 10.1186/1750-1172-5-8.
- Davis SM, Bloy L, Roberts TPL, et al. Testicular function in boys with 47,XYY and relationship to phenotype. Am J Med Genet C Semin Med Genet. 2020;184(2):371–385. doi: 10.1002/ajmg.c.31790.
- Zhang Z, Hu T, Wang J, et al. Pregnancy outcomes of fetuses with congenital heart disease after a prenatal diagnosis with chromosome microarray. Prenat Diagn. 2022;42(1):79–86. doi: 10.1002/pd.6078.
- Hu T, Tian T, Zhang Z, et al. Prenatal chromosomal microarray analysis in 2466 fetuses with ultrasonographic soft markers: a prospective cohort study. Am J Obstet Gynecol. 2021;224(5):516 e511–516 e516.
- Hu T, Zhang Z, Wang J, et al. Prenatal diagnosis of chromosomal aberrations by chromosomal microarray analysis in fetuses with ultrasound anomalies in the urinary system. Prenat Diagn. 2019;39(12):1096–1106. doi: 10.1002/pd.5550.
- Jing XY, Huang LY, Zhen L, et al. Prenatal diagnosis of 17q12 deletion syndrome: a retrospective case series. J Obstet Gynaecol. 2019;39(3):323–327. doi: 10.1080/01443615.2018.1519693.
- Zhang Z, Pan L, Chen K, et al. Prenatal ultrasound features and genetic analysis for 17q12 microdeletion syndrome. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2021;46(12):1370–1374. doi: 10.11817/j.issn.1672-7347.2021.210412.
- Khan S, Donnelly J. Outcome of pregnancy in women diagnosed with idiopathic polyhydramnios. Aust N Z J Obstet Gynaecol. 2017;57(1):57–62. doi: 10.1111/ajo.12578.
- Zeino S, Carbillon L, Pharisien I, et al. Delivery outcomes of term pregnancy complicated by idiopathic polyhydramnios. J Gynecol Obstet Hum Reprod. 2017;46(4):349–354. doi: 10.1016/j.jogoh.2017.02.014.
- Polnaszek B, Liang B, Zhang F, et al. Idiopathic polyhydramnios and neonatal morbidity at term. Am J Perinatol. 2021;40(16):1827–1833. doi: 10.1055/s-0041-1739435.
- Manickam K, McClain MR, Demmer LA, et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2021;23(11):2029–2037. doi: 10.1038/s41436-021-01242-6.