Abstract
The circadian clock controls the timing of the cell cycle in healthy tissues and clock disruption is known to increase tumourigenesis. Melanoma is one of the most rapidly increasing forms of cancer and the precise molecular circadian changes that occur in a melanoma tumor are unknown. Using a melanoma zebrafish model, we have explored the molecular changes that occur to the circadian clock within tumors. We have found disruptions in melanoma clock gene expression due to a major impairment to the light input pathway, with a parallel loss of light-dependent activation of DNA repair genes. Furthermore, the timing of mitosis in tumors is perturbed, as well as the regulation of certain key cell cycle regulators, such that cells divide arhythmically. The inability to co-ordinate DNA damage repair and cell division is likely to promote further tumourigenesis and accelerate melanoma development.
Introduction
Most organisms possess a highly conserved endogenous circadian clock, providing a clear survival advantage to animals that live under an environmental light and dark cycle.Citation1 The molecular clock mechanism operates through a transcription-translation negative feedback loop of circadian genes and proteins. The alternating activation and suppression of core clock genes, such as Clock, Bmal, Per and Cry, produce a 24 h oscillation, which can then regulate the timing of a wide range of downstream, output processes.Citation2,3 One of the most significant of these outputs is the daily control of cell proliferation and DNA repair, which has been shown in numerous tissues across many animal model systems.Citation4-7 We have previously reported that this cellular clock controls cell cycle events in normal proliferative tissues from early development until adulthood in zebrafish. This cellular clock also controls the timing of the cell cycle in cell culture, with S-phase occurring in the late day/early evening and mitosis in late night/early morning.Citation6,7 The clock regulation of key cell cycle regulators, such as the inhibitors p21 and p20, creates a window or gate that permits cells to enter S-phase from G1 when expression levels are low.Citation8
Circadian clock disruption is associated with numerous health problems and has been linked to an increased incidence of cancer.Citation9 Epidemiologic studies, for example, have revealed an increased risk of breast and colorectal cancers in night shift workers.Citation10-12 The hypothesis of a disrupted clock involved in cancer development has also been supported by studies in rodents. Mice, whose clock had been surgically disrupted by removal of the suprachiasmatic nucleus, and then inoculated with tumor cells showed an increase in tumourigenesis.Citation13 Environmental clock disruptions by constant light or jet lag exposure in rats and mice have also revealed an increase in spontaneous tumor appearance and tumor growth.Citation13-15 Several clock genes have been proposed to act as tumor suppressors and the PER family of proteins in particular appears to play a role in DNA damage repair and tumor suppression. Over expression of Per1 in colon cancer cell lines was associated with a higher level of apoptosis after irradiation, whereas inhibition of Per1 expression led to a decrease in apoptosis.Citation16 Similar effects were observed with Per2 in a leukemia cell line.Citation17 Transgenic mice lacking both Per1 and Per2 showed higher rates of tumourigenesis after irradiation.Citation18 Moreover cancer patients commonly show disruptions in their circadian clock, which is nowadays being used as a prognostic tool for breast cancer patients.Citation19 Chronochemotherapy has emerged as a treatment strategy, with the discovery of a circadian profile for drug target genes, including those involved in the cell cycle. This mode of treatment takes advantage of the asynchrony between healthy and cancerous tissues and has proven to be successful in delivering treatment at an optimal time of day to increase survival of colorectal cancer and childhood acute leukemia patients compared to normal, non-timed protocols.Citation20-22
Melanoma is a very severe and significant form of skin cancer, with approximately 20% of diagnosed individuals succumbing to the disease. It is also a form of cancer that is showing the most dramatic increase in incidence within the population. This number is set to rise even further in Western countries, as travel to sunny locations and exposure to DNA-damaging UV light has escalated in recent years.Citation23,24 Human skin is very sensitive to light exposure, especially in the UV range, which can lead to DNA damage and the initiation of melanoma.Citation25,26 Skin cells have been shown to contain a robust biological clock in mammals although the function of this skin clock is generally unknown.Citation27-30 There is evidence for a role of the circadian clock in maintaining stem cell heterogeneity in the epidermis and the timing of DNA replication appears to be under clock-control in keratinocytes.Citation31,32 Analysis of clock gene expression in human skin and melanoma tumor biopsies showed down regulation in tumor samples.Citation33 However, the circadian profile of clock genes and clock-controlled genes (CCGs) remains unexplored in melanoma. How clock-cell cycle interactions function in a melanoma tumor environment is an important issue and one of considerable clinical significance.
Zebrafish have already proven to be an excellent vertebrate system in which to study melanoma, due in part to the high gene homology to mammals in cancer related pathways.Citation34,35 Using a zebrafish melanoma model, we have analyzed the circadian profile of clock gene expression over several days in vivo and in vitro and observed a down regulation of clock gene expression in melanoma tumors compared to healthy skin. We have shown that impaired light detection in melanoma tumors may underpin the disruptions observed in central circadian clock components. It is also clear that the circadian timing of mitosis itself is disrupted in these tumors, along with corresponding changes in gene expression. Loss of light detection also compromises induction of the DNA-damage repair pathways, a fact that may promote further cellular mutations, and promote additional, accelerated tumor growth.
Results and Discussion
Clock gene expression is altered in zebrafish melanoma tumors
To explore the expression profile of clock genes in melanoma we used transgenic zebrafish Tg(mitfa:V12Ras) expressing the constitutively active V12Ras under the control of the melanocyte specific promoter mitfa.Citation35,36 These animals were crossed to nacre (mitfa−/−) animals to generate a Tg(mitfa:V12Ras);mitfa−/− strain lacking melanocytes. Injection of a miniCoopR-GFP vector [33] into zygotes from a Tg(mitfa:V12Ras);mitfa−/− × mitfa−/− cross was then used to rescue the melanocyte lineage in offspring and induce more penetrant and rapid melanoma development than observed in Tg(mitfa:V12Ras) alone. In parallel, the miniCoopR-GFP vector drives expression of GFP in rescued melanocytes.Citation36 All Tg(mitfa:V12Ras);mitfa−/−;miniCoopR-GFP+ (henceforward V12Ras+; GFP+) animals developed dysplastic melanocytic pigmentation pattern from day 4, and consequently GFP-labeled melanoma tumors at around 4 weeks of age while their mitfa−/−; miniCoopR-GFP+ (henceforward GFP+) siblings showed normal pigmentation and no tumor (Fig. S1).
Expression of key clock genes was explored in-vivo in melanoma tumors from V12Ras+; GFP+ zebrafish and compared to expression in healthy skin harvested from control GFP+ animals. qPCR analysis showed a significant down-regulation of clock gene expression in tumors compared to healthy skin samples (). Samples were collected every 6 hours over 4 days, 2 days under light-dark (LD) conditions, and 2 subsequent days in constant dark (DD), free-running conditions. In LD, we observed shallow rhythms, with significantly reduced amplitude, for the core clock genes per1, clock and bmal1a in tumor compared to robust rhythms in healthy skin (, Tables S1, S2). The significant reduction in amplitude under LD conditions is quantified in .
Figure 1. Clock gene expression is downregulated in zebrafish melanoma tumors. (A) qPCR analysis of core clock genes per1, clock1, and bmal1a in tumors and skin from animals maintained on a LD cycle, then transferred to DD. Relative expression to the reference gene represents the mean ± SEM of a minimum of 5 samples per time point. White and gray backgrounds represent light and dark phases respectively. (B) Quantification of the reduction in amplitude in tumors using skin as a reference. Data represent the mean ± SEM of 5 samples. (C) Bioluminescent traces of per3-luciferase tumors and skin in LD, then transferred to DD. Results are presented as detrended data from representative samples and white and black bars under the traces represent light and dark phases respectively. (D) Period lengths in LD and DD, calculated from the bioluminescent data, are presented in hours. (E) Amplitude differences between skin and tumors in LD and DD from the bioluminescent data are presented in counts per second (CPS). Data represent the mean ± SEM of 12 samples using a Student's t-test (unpaired, 2-tailed; ***P < 0.001).
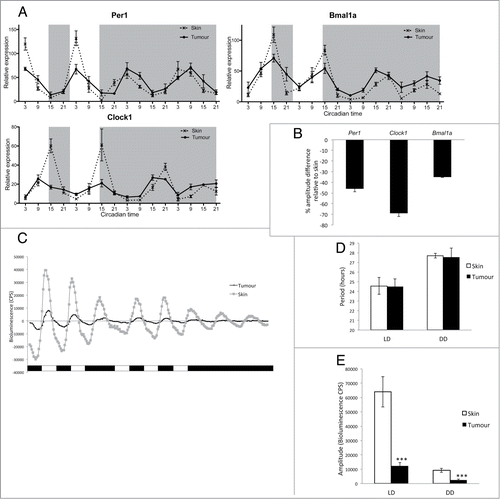
A reduction in circadian clock amplitude has been reported in other types of cancer such as breast, prostate, non-small cell lung cancer, and head and neck squamous cell carcinoma.Citation37-40 In particular, a study on human melanoma tumor biopsies has reported a significant down regulation of clock genes compared to adjacent healthy tissues.Citation33 However, human skin contains different cell types displaying a range of amplitudes in clock gene rhythms, with keratinocyte and dermal fibroblast cultures showing a more robust clock compared to melanocyte cultures.Citation28 To ensure that the difference in amplitude we observed between normal skin and melanoma tumors in our study is not due to a higher number of melanocytes in tumor samples, we analyzed clock gene expression in zebrafish dysplastic naevi (Fig. S2). Dysplastic naevi appear during the radial growth phase of melanoma development and lack alterations in certain pathways, such as phosphoinositide 3-kinase (PI3K)-AKT signaling, required for the vertical growth phase and formation of a melanoma tumor.Citation35 Naevi, therefore, provide a melanocyte-rich environment, which has not yet progressed to a malignant melanoma (Fig. S1). Analysis of per1, clock1 and bmal1a expression across one LD cycle showed no down regulation in naevus samples compared to healthy skin (Fig. S2). This result indicates that the decreased amplitude in clock gene expression seen in melanoma tumors is most likely due to malignant transformation itself and not to the abundance of melanocytes.
Interestingly, under DD conditions, we found very little disruption of clock gene expression in tumors when measured by qPCR (). However, cosinor analysis of the data revealed a significant increase of the mesor in tumors versus skin but most importantly the amplitude relative to the mesor showed a significant decrease for per1, bmal1a and clock (Tables S1, S2), suggesting a defective molecular clock in the tumor population. However, the tumor situation in vivo is quite complex in that healthy tissues are surrounding a heterogeneous tumor. To determine the impact of malignancy on clock function in the tumor more precisely it is essential to follow circadian oscillations within single tumors dynamically and in vitro. We consequently examined clock rhythms in tissue culture of single tumors and healthy skin controls expressing period3-luciferase in bioluminescent assays across 4 days in LD then transferred to DD (). Under these conditions, the period length of the circadian oscillator showed no statistical difference between tumor and skin, either in LD or DD (). However in LD, the amplitude was dramatically reduced in tumors compared to skin (), even more so than in our in vivo qPCR analysis (). Moreover, the amplitude of tumor circadian rhythms dampens rapidly when transferred to DD compared to data shown in vivo by qPCR in (). The circadian pacemaker in tumors in vitro, therefore, appears more disrupted than in vivo, possibly due to the lack of support from surrounding healthy tissue. This suggests that healthy surrounding tissues are likely to play a role in maintaining tumor rhythmicity in vivo, though the mechanism of oscillator coupling is far from clear. Nevertheless, we have shown that the circadian clock does continue to function within the melanoma, if with reduced amplitude. This reduction in robustness could be due to a variety of alterations in tumor clock function, including perturbation of the light input pathway, reduced cellular coupling between oscillators or disruption in the regulation of the core clock mechanism within each cell.
Impaired light detection in the tumor reduces induction of genes involved in clock entrainment and the DNA-damage repair system
Light is the main entraining signal to the circadian clock, and the majority of zebrafish cells and tissues are themselves directly light responsive.Citation41 Two light-inducible clock genes, cry1a and per2, have been shown to play a critical role on the light input pathway to the clock in zebrafish.Citation42,43 We examined by qPCR the expression of these 2 genes in vivo in LD and then transferred into DD. Cry1a expression was dramatically down regulated in tumor samples in LD, but showed similar levels in DD compared to skin samples (). Cry1a expression is under clock control as well as being light inducible, a fact that explains the similar oscillation that continues in constant darkness. The light-inducible clock gene most disrupted in the tumor was per2 with more than an 80% reduction in amplitude in tumor samples compared to skin (, Fig. S3). The dramatic reduction in expression of the 2 genes known to be involved in zebrafish clock entrainment is likely to contribute to the reduced amplitude rhythms in LD shown in . In many respects, it is an unexpected result that the light input pathway to the clock is so strongly affected in this zebrafish model of melanoma, even when one takes into account the fact that the skin is very strongly light responsive. In mammals there is no evidence at this time that peripheral tissues are themselves directly light sensitive. A detailed analysis of clock function in a mammalian melanoma context is certainly important. However, many of the genes involved in the clock system are highly conserved between zebrafish and mammals, even if there are changes in their specific roles. This is certainly true in the case of both period 2 and cryptochrome genes. There is also considerable evidence that per2 can act as a tumor suppressor in mammalian systems.Citation44,45 Disruption of per2 expression, either when it is acting as a component of the circadian clock mechanism as in mammals, or the light input pathway as in zebrafish, could potentially play a significant role in enhanced tumourigenesis.
Figure 2. Impaired light detection and DNA-damage repair system. (A) qPCR analysis of light inducible clock genes per2 and cry1a and light dependent DNA-damage repair genes 6,4ph (6,4 photolyase) and ddb2 (DNA damage binding protein 2). Relative expression to the reference gene represents the mean ± SEM of minimum 5 samples per time point. (B) qPCR analysis cry1a, per2, ddb2 and 6,4ph expression in skin and tumor after a 3 hour light pulse given at CT16 compared to samples kept in the dark (DD). Data represent the mean ± SEM of a minimum of 5 samples per time point. (C) Absolute fold induction of each gene in response to light in skin and tumor. Fold induction was compared between skin and tumor using a Student's t-test (unpaired, 2 tailed; **P < 0.01; ***P < 0.001). Data represent the mean ± SEM of 8 samples.
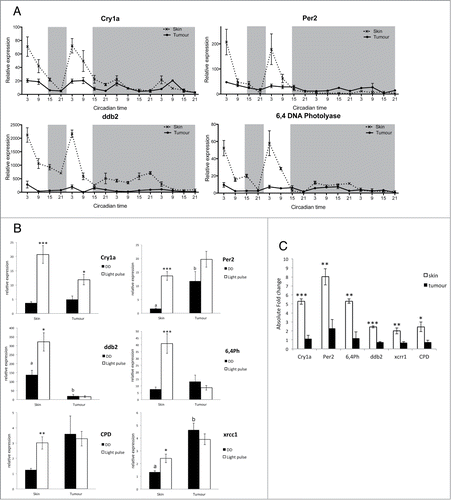
Melanoma tumors often appear black due to the melanin pigment present in melanocytes. To ensure that this extra pigmentation of the tissue does not affect the ability of the tumor to perceive light, we analyzed the expression of per2 and cry1a in naevi, which are also highly pigmented. Per2 and cry1a showed no down regulation of expression in naevi but interestingly a slightly greater amplitude compared to skin samples (Fig. S4). We can therefore conclude that there is a defect in the pathway leading to the induction of light responsive genes in melanoma tumors, not resulting from the increased pigmentation, and which may lead to a reduction in the robustness of the clock rhythm measured in these tumors relative to neighboring, healthy tissue.
Transcriptional regulation by light can also directly affect other cellular processes, such as DNA damage repair pathways.Citation46,47 Expression of the DNA-damage repair genes ddb2 and 6,4 photolyase (6,4ph) displays a robust oscillation in LD peaking 3 h after lights on (Zeitgeber Time—ZT3) in skin samples, which is then lost when tissues are transferred to DD (). However, the expression profile of these genes in tumor samples lacks such oscillations in LD, showing arrhythmic expression and a reduction of more than 80% in expression levels compared to skin samples (, Fig. S3). A cosinor analysis of these data confirms that there are no significant rhythms in tumor samples (Tables S3, S4), and shows that the loss of light sensitivity impacts not only clock gene expression, but also DNA repair gene induction even more dramatically. Analysis of ddb2 and 6,4ph gene expression across a LD cycle in naevi compared to tumor and skin samples showed a gradient of down regulation (Fig. S5). This is in agreement with studies on dysplastic melanocytes displaying an impaired DNA damage repair system, creating an environment prone to increased genome instability and allowing for the naevi to progress to melanoma.Citation48 This partial loss of light induction in DNA damage repair genes in naevi, at an early stage of melanoma development, establishes a situation that promotes and possibly even accelerates subsequent tumor growth.
To assess the light input pathway in melanoma tumors, we examined the induction of cry1a, per2, and 4 DNA damage repair genes following a 3 h light pulse during the subjective night (). The expression of all genes was greatly enhanced following the light pulse in control skin samples, as previously reported in zebrafish cell lines and larvae.Citation42,47 The acute light response was significantly reduced for cry1a expression in tumors, and no significant response occurred at all for per2 and DNA-damage repair genes (). All light-inducible genes examined showed a significant absolute fold reduction in expression, which demonstrates the profound impairment in the light detection pathway in melanoma tumors (). A functional DNA repair system is essential to protect against mutations caused by UVA and UVB light, especially in skin cells. ddb2 and 6,4ph expression in healthy skin was shown to peak at the start of the day therefore providing optimal protection against UV-induced mutations. Melanoma tumors, however, lack light inducible expression of these key DNA damage repair genes, which suggest that their DNA-repair mechanisms will be compromised, and consequently, they are prone to attaining even greater levels of DNA damage and genomic instability.
Melanoma tumors display disruption in the timing of clock output events
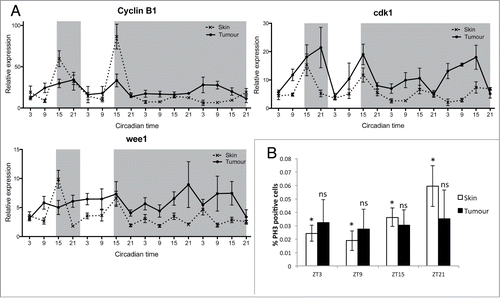
Controlled timing of the cell cycle is one of the major circadian clock outputs. We have previously reported that key cell cycle regulators are clock controlled in zebrafish cell lines and embryos.Citation6,8,49 To explore the circadian expression profiles of cell cycle genes in healthy skin and melanoma tumors, we collected tissue samples at 6-hour intervals over LD and DD cycles for 4 days. qPCR analysis of mitotic-related genes (cyclin B1, cdk1, wee1) in skin showed rhythmic expression in LD, with peak expression during the early night (). In comparison, expression in tumor showed clear disruption, including loss of rhythmicity in wee1 in LD, and amplitude variations in cyclinB1 and cdk1. All mitotic genes showed a loss of rhythmicity in DD (, Tables S5, S6), suggesting that the circadian clock might have reduced control over the timing of mitosis. To determine if this disruption in mitosis-related gene expression affected actual mitotic timing, we used a phospho-histone H3 (pH3) antibody to label cells in mitosis in zebrafish skin and tumor sections at 4 different time points across a LD cycle. Skin samples showed a rhythm in mitosis with the highest percentage of pH3 positive cells found during the late night (ZT21) (). Tumor samples did not show any rhythm in mitotic events with a high degree of variability in the percentage of pH3 positive cells at each time point. Interestingly, the average index of mitotic events remained similar throughout the skin and tumor samples, suggesting that the overall number of cells undergoing mitosis is very similar (Fig. S6). Most healthy skin cells undergo mitosis at night, possibly to avoid any negative consequences caused by UV light exposure during the day, whereas tumor cells divide randomly at any time of day.
Figure 3. Disruption in mitotic events in melanoma tumors. (A) qPCR analysis of mitotic genes cyclin B1, cdk1 and wee1 in healthy skin and tumors. Relative expression to the reference gene represents the mean ± SEM of minimum 5 samples per time point. (B) Quantification of mitotic events in healthy skin and tumors using an antibody to phospho-Histone H3 (pH3). Data represent the percentage of pH3-positive cells relative to the total number of cells in one section. Data represent the mean ± SEM of a minimum of 3 different fish. The percentage of pH3 positive cells were compared at each time point for skin and tumor using a one-way ANOVA test followed by a Newman-Keuls multiple comparison post test (*P < 0.05, ns = non significant).
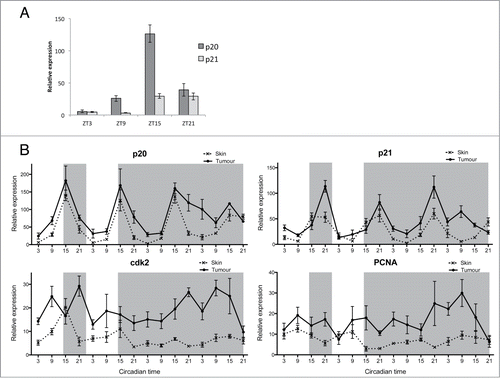
We have hypothesized that the timing of S-phase entry is regulated by 2 CCGs, p20 and p21, which control tissue specific S-phase timing in zebrafish.Citation8 To identify which of these inhibitors is the most abundant in skin we compared their expression across a LD cycle in skin samples and found that p20 is significantly more abundant than p21 in this particular tissue (). The circadian profile of p20 expression showed robust rhythms in skin samples, peaking in the night at ZT15 (). We have previously reported similar findings in zebrafish embryos, where p20 is the main regulator in the developing brain. In the developing zebrafish larvae p20 shows a clear phase difference in expression compared to p21, where it is responsible for timing actual S-phase events to ZT3, in the early day.Citation8 This peak at ZT3 in S-phase would be highly appropriate in skin, as it would coincide with the peak in expression of DNA repair genes, therefore possibly providing protection against UV light-induced mutations during the process of DNA replication. Surprisingly, tumor samples revealed a similar p20 expression pattern in LD, suggesting that the regulation of S-phase timing is not disrupted in tumors unlike the regulation of mitosis. However, rhythms in p20 in DD do become less precise over time in tumors, which may be a reflection of the somewhat disrupted circadian pacemaker in these cells. Other S-phase genes such as p21, cdk2 and PCNA showed an up-regulation in expression in tumor samples in LD and DD, with a loss of rhythmicity in DD for cdk2 and PCNA compared to skin samples (, Tables S5, S6). We can conclude that the cell cycle in melanoma tumors displays disrupted circadian properties with a loss in rhythmicity of several genes, especially those involved in mitosis.
Figure 4. Disruption in S-phase gene expression in melanoma tumor. (A) qPCR analysis of S-phase regulating genes p21, cdk2, and PCNA in skin and tumor. Relative expression to the reference gene represents the mean ± SEM of a minimum of 5 samples per time point. (B) Cosinor analysis from qPCR values presented in (A). Data represent the mesor and amplitude in relative expression, the acrophase in circadian time and the significance of rhythmicity (*P < 0.05; ***P < 0.001) for skin and tumor in LD and DD regimes.
We have demonstrated in this study that light-inducible genes, such as cry1a and per2, showed a reduced response to light in zebrafish melanoma compared to healthy skin. This impaired light response also resulted in a total loss in expression of light-induced DNA damage repair genes during the daytime, when UV-induced mutation risk is at its highest. We believe that this impaired light response, observed in tumors, could reduce the strength of entrainment of the clock and may lead to the reduced amplitude in clock gene oscillations in LD. The lower level of expression of clock genes in tumors is also likely to reduce the level of precise control over cellular clock outputs, such as the cell cycle. Key cell cycle genes show a loss of circadian regulation, and mitotic events consequently are also no longer rhythmic, meaning that cell division occurs to a greater extent during the day compared to healthy skin. The coupling process between the circadian clock and control of mitosis is clearly disrupted in melanoma. Loosing the ability to repair DNA combined with incorrect circadian timing of cell cycle events is likely to create a highly mutagenic environment for cells in the tumor and promote tumourigenesis even further. Genomic instability and sustained proliferative signaling are cancer hallmarks.Citation50 We therefore identified a crucial role for the zebrafish skin clock, which is to time the cell cycle to periods of darkness or times of highest active DNA repair in order to confer protection from UV in dividing cells.
What might be the cause of this decoupling between the cellular clock and the timing of mitotic events within the melanoma? Light is typically believed to be the dominant entraining signal for the circadian clock. However, glucocorticoid signaling has also been shown to play a key role in entrainment, especially of peripheral circadian pacemakers.Citation51 Moreover, the presence of glucocorticoid signaling/cortisol has been shown to be important for the control of rhythmic cell cycle events in healthy tissues during zebrafish embryo development.Citation52 In the case of the strong rx3 zebrafish mutants, which possess fewer corticotropes within the anterior pituitary and reduced levels of cortisol, the molecular circadian clock appears to be normal, but there is a strong reduction in the amplitude of the circadian rhythm in S-phase, as measured by BrdU incorporation into the skin. Tonic treatment of strong rx3 mutant larvae with the glucocorticoid receptor agonist, dexamethasone, can largely rescue this cell cycle rhythm, re-establishing the coupling between clock and cell cycle. These results have some clear similarities with the data we have obtained in melanoma. Though the clock in the tumor is somewhat more disrupted, especially in terms of the light input pathway, it is still able to show clear daily molecular oscillations, yet it is the coupling to control mitotic rhythms that is clearly lost. This raises the interesting possibility that there maybe a disruption in glucocorticoid signaling in the context of the melanoma tumor. This, of course, could occur at many levels from disruption of glucocorticoid receptors in the tumor, through to perturbation of cortisol levels in an unhealthy animal. Clearly, future studies will need to explore the possible role played by glucocorticoid signaling in coupling clock and cell cycle, especially during the process of tumourigenesis. Zebrafish melanoma certainly offers a unique model in which to study the intricate link between the circadian clock, signaling pathways and the cell cycle.
Revealing how clock-cell cycle interactions function in a cancer environment is an important issue, not only to understand the circadian biology of cancer, but also to provide insights on how to improve cancer treatment through chronotherapy. Chronotherapeutic regimes, delivering drugs at specific times of the day for optimal drug action and reduced toxic side effects, have already shown promising results in metastatic prostate cancer patients in a timed regime compared to conventional treatment.Citation21 The circadian profile of cell cycle genes generated in this study clearly points to an asynchrony in melanoma tumors compared to healthy skin cells. In such a scenario, the potential exists, therefore, to apply drugs to kill cancer cells at a time where there will be minimal impact on healthy tissue. Zebrafish have already been described as an excellent model for pharmacological studies and future experiments will assess the efficacy of key melanoma drugs within a chronochemotherapeutic regime.
Materials and Methods
Animal husbandry
Tg(mitfa:V12Ras); mitfa−/− animals were obtained from Dr. Adam Hurlstone (University of Manchester) and were raised and maintained in the zebrafish animal facility of University College London, as previously described.Citation53 All animals were held in a Home Office approved animal facility and in accordance with Home Office regulations regarding animal maintenance and care. Animal handling has been approved by a UCL ethics committee and meets all of the requirements of the Animal Welfare Act of 2006. Individual experiments were performed under animal license number PIL 40/3292. Animals were sacrificed in accordance with Schedule 1 of the Animal Welfare Act of 2006, to ensure minimal suffering. Adult fish were kept in light cabinets and exposed to a lighting regime of 14L/10D unless stated otherwise.
Generation of transgenic melanoma fish
The miniCoopR GFP vector was a gift from Adam Hurlstone (University of Manchester).Citation36 75pg of miniCoopR-GFP vector and 75pg of tol2 transposase mRNA were microinjected into one-cell stage embryos generated from a (mitfa:RasV12); mitfa−/− × mitfa−/− zebrafish cross. Transgenic animals recapitulated a melanoma tumor phenotype observed in the Tg(mitfa:RasV12),Citation35 but with an earlier onset of 4 weeks of age and with far greater penetrance as all fish developed tumors. Animals developing either melanoma tumors or normal pigmentation were used for tumor and skin sample collection, respectively. It should be noted that the tumor bearing fish are culled before the size of the tumor affect their feeding and swimming, so the presence of the tumor does not affect fish survival.
Quantitative PCR (qPCR)
Adult skin and tumor samples were harvested at the indicated zeitgeber or circadian time (ZT or CT, where ZT0 equals lights on). RNA extraction, cDNA synthesis and qPCR experiments were carried out as previously described.Citation8 ΔCT was calculated using ribosomal 18S as a reference gene. Relative expression levels were plotted after determining ΔΔCt by normalizing to a single sample with a high ΔCT value. Primer sequences are listed in Table S7.
Bioluminescence assays
Skin and tumor samples from V12Ras+; GFP+ and GFP+ injected animals in a per3-luciferase background were dissected and placed in medium containing 0.5mM of luciferin (Promega) in a 96-well plate. Samples were maintained at 28°C on a light-dark (LD) cycle (12L:12D) and transferred into either constant darkness (DD) or a reverse light/dark cycle. Bioluminescence was monitored on a Packard TopCount NXTscintillation counter. The luminescent rhythm parameters (phase and amplitude) were calculated after detrending by subtracting a 24-h moving average from the raw data.
Phospho-Histone H3 immunohistochemistry
Whole zebrafish were fixed at specified time points on a LD cycleCitation33 and sectioned as previously describedCitation49 with the following changes. Animals were fixed in 4% PFA in 0.1 phosphate buffer at 4°C for 5 days, then transferred to a 0.25 M EDTA solution for 3 days at room temperature. EDTA was rinsed off with water before immersing the samples in 30% sucrose solution for an additional 2 days. Tumor-bearing and healthy fish were cryosectioned at 10μm and stained, as previously described.Citation49 Images were collected using a Zeiss AxioScan Z1 slide scanner. Quantification of pH3-positive cells was calculated relative to the total number of cell nuclei in skin layers, stained with DAPI and within the GFP-labeled tumor area.
Statistical analysis
The data in this study are presented as the mean ± SEM (n ≥ 3). Statistical significance was determined by an unpaired 2-tailed Student t-test or analysis of variance (ANOVA), followed by Newman–Keuls multiple comparison post-test in GraphPad Prism.
Light pulse experiments
Fish were kept on a LD cycle for 7 days before being transferred to DD. At CT16 on the first DD night, animals were exposed to a 3 h light pulse using LED lights with an average intensity of 65 μW/cm2.
Rhythm analysis
Rhythm analyses were performed by the Cosinor method (Nelson et al. 1979), using the El Temps software developed by Prof. A. Díez Noguera. Rhythms are considered significant when P < 0.05. *P < 0.05; **P < 0.01; ***P < 0.001 and non-significant rhythms when P < 0.1.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
NH and DW designed research and wrote the paper, and NH and ND performed research.
Acknowledgements
We thank Dr Agueda Jimena Martin Robles and Prof. Díez-Noguera for help using the El Temps software and the UCL zebrafish facility staff for their assistance. We thank Dr Adam Hurlstone and Dr Masazumi Tada for critically reading the manuscript and Dr Adam Hurlstone for supplying zebrafish lines and other reagents.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
Funding was kindly provided by the BBSRC to David Whitmore—BB/I003592/1.
References
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 2005; 6:544-56; PMID:15951747; http://dx.doi.org/10.1038/nrg1633
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet 2008; 9:764-75; PMID:18802415; http://dx.doi.org/10.1038/nrg2430
- Kondratov RV, Gorbacheva VY, Antoch MP. The role of mammalian circadian proteins in normal physiology and genotoxic stress responses. Curr Top Dev Biol 2007; 78:173-216; PMID:17338917; http://dx.doi.org/10.1016/S0070-2153(06)78005-X
- Johnson CH. Circadian clocks and cell division: what's the pacemaker? Cell Cycle 2010; 9:3864-73; PMID:20890114; http://dx.doi.org/10.4161/cc.9.19.13205
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science 2003; 302:255-9; PMID:12934012; http://dx.doi.org/10.1126/science.1086271
- Dekens MPS, Santoriello C, Vallone D, Grassi G, Whitmore D, Foulkes NS. Light regulates the cell cycle in zebrafish. Curr Biol 2003; 13:2051-7; PMID:14653994; http://dx.doi.org/10.1016/j.cub.2003.10.022
- Tamai TK, Young LC, Cox CA, Whitmore D. Light acts on the zebrafish circadian clock to suppress rhythmic mitosis and cell proliferation. J Biol Rhythms 2012; 27:226-36; PMID:22653891; http://dx.doi.org/10.1177/0748730412440861
- Laranjeiro R, Tamai TK, Peyric E, Krusche P, Ott S, Whitmore D. Cyclin-dependent kinase inhibitor p20 controls circadian cell-cycle timing. Proc Natl Acad Sci U S A 2013; 110:6835-40; PMID:23569261; http://dx.doi.org/10.1073/pnas.1217912110
- Sahar S, Sassone-Corsi P. Circadian clock and breast cancer: a molecular link. Cell Cycle 2007; 6:1329-31; PMID:17534151; http://dx.doi.org/10.4161/cc.6.11.4295
- Hansen J. Light at night, shiftwork, and breast cancer risk. J Natl Cancer Inst 2001; 93:1513-5; PMID:11604468; http://dx.doi.org/10.1093/jnci/93.20.1513
- Davis S, Mirick DK, Stevens RG. Night shift work, light at night, and risk of breast cancer. J Natl Cancer Inst 2001; 93:1557-62; PMID:11604479; http://dx.doi.org/10.1093/jnci/93.20.1557
- Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, Fuchs CS, Colditz GA. Night-shift work and risk of colorectal cancer in the nurses' health study. J Natl Cancer Inst 2003; 95:825-8; PMID:12783938; http://dx.doi.org/10.1093/jnci/95.11.825
- Filipski E, Levi F. Circadian disruption in experimental cancer processes. Integr Cancer Ther 2009; 8:298-302; PMID:20042408; http://dx.doi.org/10.1177/1534735409352085
- Vinogradova IA, Anisimov VN, Bukalev AV, Semenchenko AV, Zabezhinski MA. Circadian Disruption Induced by Light-at-Night Accelerates Aging and Promotes Tumorigenesis in Rats. Aging (Albany NY) 2009; 1:855-65.
- Filipski E, Delaunay F, King VM, Wu MW, Claustrat B, Grechez-Cassiau A, Guettier C, Hastings MH, Francis L. Effects of chronic jet lag on tumor progression in mice. Cancer Res 2004; 64:7879-85; PMID:15520194; http://dx.doi.org/10.1158/0008-5472.CAN-04-0674
- Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell 2006; 22:375-82; PMID:16678109; http://dx.doi.org/10.1016/j.molcel.2006.03.038
- Sun CM, Huang SF, Zeng JM, Liu DB, Xiao Q, Tian WJ, Zhu XD, Huang ZG, Feng WL. Per2 inhibits k562 leukemia cell growth in vitro and in vivo through cell cycle arrest and apoptosis induction. Pathol Oncol Res 2010; 16:403-11; PMID:19957060; http://dx.doi.org/10.1007/s12253-009-9227-0
- Lee S, Donehower LA, Herron AJ, Moore DD, Fu L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One 2010; 5:e10995; PMID:20539819; http://dx.doi.org/10.1371/journal.pone.0010995
- Mormont MC, Levi F. Circadian-system alterations during cancer processes: a review. Int J Cancer 1997; 70:241-7; PMID:9009166; http://dx.doi.org/10.1002/(SICI)1097-0215(19970117)70:2%3c241::AID-IJC16%3e3.0.CO;2-L
- Levi F. From circadian rhythms to cancer chronotherapeutics. Chronobiol Int 2002; 19:1-19; PMID:11962669; http://dx.doi.org/10.1081/CBI-120002676
- Levi F, Zidani R, Misset JL. Randomised multicentre trial of chronotherapy with oxaliplatin, fluorouracil, and folinic acid in metastatic colorectal cancer. International Organization for Cancer Chronotherapy. Lancet 1997; 350:681-6; PMID:9291901; http://dx.doi.org/10.1016/S0140-6736(97)03358-8
- Rivard GE, Infante-Rivard C, Hoyoux C, Champagne J. Maintenance chemotherapy for childhood acute lymphoblastic leukaemia: better in the evening. Lancet 1985; 2:1264-6; PMID:2866334; http://dx.doi.org/10.1016/S0140-6736(85)91551-X
- de Vries E, Coebergh JW. Cutaneous malignant melanoma in Europe. Eur J Cancer 2004; 40:2355-66; PMID:15519506; http://dx.doi.org/10.1016/j.ejca.2004.06.003
- de Vries E, Coebergh JW. Melanoma incidence has risen in Europe. Bmj 2005; 331:698; PMID:16179724; http://dx.doi.org/10.1136/bmj.331.7518.698
- Parkin DM, Mesher D, Sasieni P. 13. Cancers attributable to solar (ultraviolet) radiation exposure in the UK in 2010. Br J Cancer 2011; 105 Suppl 2:S66-9; PMID:22158324; http://dx.doi.org/10.1038/bjc.2011.486
- Brash DE. Sunlight and the onset of skin cancer. Trends Genet 1997; 13:410-4; PMID:9351343; http://dx.doi.org/10.1016/S0168-9525(97)01246-8
- Lengyel Z, Battyani Z, Szekeres G, Csernus V, Nagy AD. Circadian clocks and tumor biology: what is to learn from human skin biopsies? Gen Comp Endocrinol 2013; 188:67-74; PMID:23608545; http://dx.doi.org/10.1016/j.ygcen.2013.03.033
- Sandu C, Dumas M, Malan A, Sambakhe D, Marteau C, Nizard C, Schnebert S, Perrier E, Challet E, Pévet P, et al. Human skin keratinocytes, melanocytes, and fibroblasts contain distinct circadian clock machineries. Cell Mol Life Sci 2012; 69:3329-39; PMID:22627494; http://dx.doi.org/10.1007/s00018-012-1026-1
- Bjarnason GA, Jordan RC, Wood PA, Li Q, Lincoln DW, Sothern RB, Hrushesky WJ, Ben-David Y. Circadian expression of clock genes in human oral mucosa and skin: association with specific cell-cycle phases. Am J Pathol 2001; 158:1793-801; PMID:11337377; http://dx.doi.org/10.1016/S0002-9440(10)64135-1
- Kawara S, Mydlarski R, Mamelak AJ, Freed I, Wang B, Watanabe H, Shivji G, Tavadia SK, Suzuki H, Bjarnason GA, et al. Low-dose ultraviolet B rays alter the mRNA expression of the circadian clock genes in cultured human keratinocytes. J Invest Dermatol 2002; 119:1220-3; PMID:12485420; http://dx.doi.org/10.1046/j.1523-1747.2002.19619.x
- Janich P, Pascual G, Merlos-Suarez A, Batlle E, Ripperger J, Albrecht U, Cheng HY, Obrietan K, Di Croce L, Benitah SA. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature 2011; 480:209-14; PMID:22080954; http://dx.doi.org/10.1038/nature10649
- Geyfman M, Kumar V, Liu Q, Ruiz R, Gordon W, Espitia F, Cam E, Millar SE, Smyth P, Ihler A, et al. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc Natl Acad Sci U S A 2012; 109:11758-63; PMID:22753467; http://dx.doi.org/10.1073/pnas.1209592109
- Lengyel Z, Lovig C, Kommedal S, Keszthelyi R, Szekeres G, Battyani Z, Csernus V, Nagy AD. Altered expression patterns of clock gene mRNAs and clock proteins in human skin tumors. Tumour Biol 2013; 34:811-9; PMID:23242607; http://dx.doi.org/10.1007/s13277-012-0611-0
- White R, Rose K, Zon L. Zebrafish cancer: the state of the art and the path forward. Nat Rev Cancer 2013; 13:624-36; PMID:23969693; http://dx.doi.org/10.1038/nrc3589
- Michailidou C, Jones M, Walker P, Kamarashev J, Kelly A, Hurlstone AF. Dissecting the roles of Raf- and PI3K-signalling pathways in melanoma formation and progression in a zebrafish model. Dis Models Mechan 2009; 2:399-411; PMID:19470611; http://dx.doi.org/10.1242/dmm.001149
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferré F. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 2011; 471:513-7; PMID:21430779; http://dx.doi.org/10.1038/nature09806
- Cao Q, Gery S, Dashti A, Yin D, Zhou Y, Gu J, Koeffler HP. A role for the clock gene per1 in prostate cancer. Cancer Res 2009; 69:7619-25; PMID:19752089; http://dx.doi.org/10.1158/0008-5472.CAN-08-4199
- Winter SL, Bosnoyan-Collins L, Pinnaduwage D, Andrulis IL. Expression of the circadian clock genes Per1 and Per2 in sporadic and familial breast tumors. Neoplasia 2007; 9:797-800; PMID:17971899; http://dx.doi.org/10.1593/neo.07595
- Hsu CM, Lin SF, Lu CT, Lin PM, Yang MY. Altered expression of circadian clock genes in head and neck squamous cell carcinoma. Tumour Biol 2012; 33:149-55; PMID:22081375; http://dx.doi.org/10.1007/s13277-011-0258-2
- Gery S, Komatsu N, Kawamata N, Miller CW, Desmond J, Virk RK, Marchevsky A, Mckenna R, Taguchi H, Koeffler HP. Epigenetic silencing of the candidate tumor suppressor gene Per1 in non-small cell lung cancer. Clin Cancer Res 2007; 13:1399-404; PMID:17332281; http://dx.doi.org/10.1158/1078-0432.CCR-06-1730
- Whitmore D, Foulkes NS, Sassone-Corsi P. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature 2000; 404:87-91; PMID:10716448; http://dx.doi.org/10.1038/35003589
- Tamai TK, Young LC, Whitmore D. Light signaling to the zebrafish circadian clock by Cryptochrome 1a. Proc Natl Acad Sci USA 2007; 104:14712-7; PMID:17785416; http://dx.doi.org/10.1073/pnas.0704588104
- Ziv L, Levkovitz S, Toyama R, Falcon J, Gothilf Y. Functional development of the zebrafish pineal gland: light-induced expression of period2 is required for onset of the circadian clock. J Neuroendocrinol 2005; 17:314-20; PMID:15869567; http://dx.doi.org/10.1111/j.1365-2826.2005.01315.x
- Hwang-Verslues WW, Chang PH, Jeng YM, Kuo WH, Chiang PH, Chang YC, Hsieh TH, Su FY, Lin LC, Abbondante S, et al. Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc Natl Acad Sci U S A 2013; 110:12331-6; PMID:23836662; http://dx.doi.org/10.1073/pnas.1222684110
- Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 2002; 111:41-50; PMID:12372299; http://dx.doi.org/10.1016/S0092-8674(02)00961-3
- Weger BD, Sahinbas M, Otto GW, Mracek P, Armant O, Dolle D, Lahiri K, Vallone D, Ettwiller L, Geisler R, et al. The light responsive transcriptome of the zebrafish: function and regulation. PLoS One 2011; 6:e17080; PMID:21390203; http://dx.doi.org/10.1371/journal.pone.0017080
- Tamai TK, Vardhanabhuti V, Foulkes NS, Whitmore D. Early embryonic light detection improves survival. Curr Biol 2004; 14:R104-5; PMID:14986634; http://dx.doi.org/10.1016/j.cub.2004.01.014
- Elder DE. Dysplastic naevi: an update. Histopathology 2010; 56:112-20; PMID:20055909; http://dx.doi.org/10.1111/j.1365-2559.2009.03450.x
- Peyric E, Moore HA, Whitmore D. Circadian clock regulation of the cell cycle in the zebrafish intestine. PLoS One 2013; 8:e73209; PMID:24013905; http://dx.doi.org/10.1371/journal.pone.0073209
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/10.1016/j.cell.2011.02.013
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schütz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 2000; 289:2344-7; PMID:11009419; http://dx.doi.org/10.1126/science.289.5488.2344
- Dickmeis T, Lahiri K, Nica G, Vallone D, Santoriello C, Neumann CJ, Hammerschmidt M, Foulkes NS. Glucocorticoids play a key role in circadian cell cycle rhythms. PLoS Biol 2007; 5:e78; PMID:17373855; http://dx.doi.org/10.1371/journal.pbio.0050078
- Westerfield M. The Zebrafish Book. A Guide for the Laboratory use of Zebrafish (Danio Rerio). Eugene: Institute of Neuroscience, University of Oregon, 1993.
