ABSTRACT
The pluripotency of newly developed human induced pluripotent stem cells (iPSCs) is usually characterized by physiological parameters; i.e., by their ability to maintain the undifferentiated state and to differentiate into derivatives of the 3 germ layers. Nevertheless, a molecular comparison of physiologically normal iPSCs to the “gold standard” of pluripotency, embryonic stem cells (ESCs), often reveals a set of genes with different expression and/or methylation patterns in iPSCs and ESCs. To evaluate the contribution of the reprogramming process, parental cell type, and fortuity in the signature of human iPSCs, we developed a complete isogenic reprogramming system. We performed a genome-wide comparison of the transcriptome and the methylome of human isogenic ESCs, 3 types of ESC-derived somatic cells (fibroblasts, retinal pigment epithelium and neural cells), and 3 pairs of iPSC lines derived from these somatic cells. Our analysis revealed a high input of stochasticity in the iPSC signature that does not retain specific traces of the parental cell type and reprogramming process. We showed that 5 iPSC clones are sufficient to find with 95% confidence at least one iPSC clone indistinguishable from their hypothetical isogenic ESC line. Additionally, on the basis of a small set of genes that are characteristic of all iPSC lines and isogenic ESCs, we formulated an approach of “the best iPSC line” selection and confirmed it on an independent dataset.
Introduction
Human pluripotent stem cell (PSC) lines can be cultured and indefinitely expanded in vitro without loss of their capacity to differentiate into a variety of cell types. There are 2 types of human PSCs: embryonic stem cells (ESCs) and induced (i) PSCs. The former were first established in 1998,Citation1 and their differentiated derivatives are now in clinical trials for allogeneic cell replacement therapy.Citation2,3 iPSCs are generated by somatic cell reprogramming and, despite minor differences, are quite similar to ESCs in their functional and molecular properties.Citation4-8 Because they are patient-specific, iPSC lines can be used in a wide range of biomedical applications.Citation9-11 However, the extent of the similarity between iPSCs and the “gold standard” of pluripotency, human ESCs, is still unclear. Indeed, the tetraploid complementation approach can be used to determine this similarity for mouse iPSCs; however, it is not applicable to humans and other species. Several groups have already identified epigenetic and gene expression signatures specific to iPSCs, as well as hot spots for aberrant methylation and somatic memory retention in mouse and human iPSCs.Citation6,8,12-15 These studies highlighted significant differences between iPSCs and ESCs, although only a limited number of cell lines of different origins were analyzed. Thus, individual genome characteristics impact cell line diversity. Later, a comprehensive characterization of dozens of human PSC lines was performed,Citation4,16 demonstrating that as more cell lines are taken into analysis, fewer differences are observed.Citation17 Recently, an effective tool to validate self-renewal potential, as well as differentiated states of iPSC lines with diverse genetic backgrounds, has been developed.Citation4 However, the need to differentiate a particular iPSC line into multiple lineages; i.e., in the case of banked HLA homozygous cells, ultimately raises the issue of iPSCs quality in respect to their genotype-specific pluripotent state and similarity to preexisting ESCs. Multiplication of the cell lines in the studies provides a better overview of the accuracy of reprogramming on average, but does not determine whether an iPSC line chosen for multiple applications corresponds to its predecessor ESC and mimics all of its properties necessary for establishing an accurate genotype-specific status of pluripotency. The only way to determine if somatic cells have returned to their initial pluripotent state is to compare iPSCs to the isogenic ESC line.
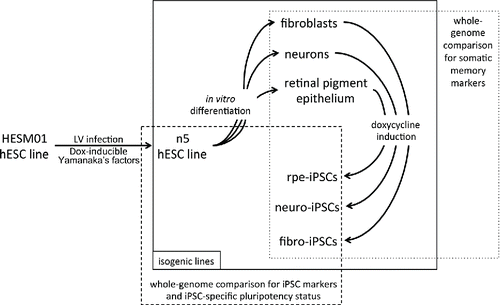
To obtain comprehensive data on the transcriptional and epigenetic variations that are gained during the reprogramming process, we compared iPSC lines generated from different somatic cell types that have been previously differentiated from ESCs. Reprogramming factors under the control of doxycycline (DOX)-inducible promoters were introduced into hESCs. Standard differentiation protocols and separation methods were used to obtain pure populations of several somatic cell types, which were further reprogrammed by adding DOX ().
Figure 1. Schematic representation of the experimental procedure. Lentiviral vectors carrying reprogramming factors were introduced into the hESM01 cell line, and the stable clones were selected for further analysis (zero transgene expression, genome stability, in vitro and in vivo pluripotency). The resulting hESM01-OSKMN-DOX-n5 cell line was differentiated into 3 types of somatic cells. Magnetically separated cells were reprogrammed by DOX induction and iPSC clones generated from each cell type were chosen for genome-wide analysis of reprogramming traces, somatic memory, and iPSC specific markers using transcription and DNA methylation data.
We performed 2 genome-wide assays to analyze the methylation and expression patterns of 11 isogenic human cell lines, including 8 PSC and 3 somatic cell lines. We showed that the reprogramming process itself and the parental somatic cell type did not leave any specific signature in iPSCs; that is, the observed differences between hESCs and isogenic iPSCs were specific to a particular clone but not to the process or predecessor cells. Because no common iPSC specific signature has been observed even for a single batch of isogenic lines, it is likely that none exists for other isogenic clones or non-isogenic lines. Additionally, variability between isogenic iPSCs derived from different somatic cell types allowed us to propose an approach for finding the optimal iPSC clone (i.e., the one most closely resembling its hypothetical isogenic human ESC line) in the cohort.
Results
Establishment of the hESM01-OSKMN-DOX isogenic system
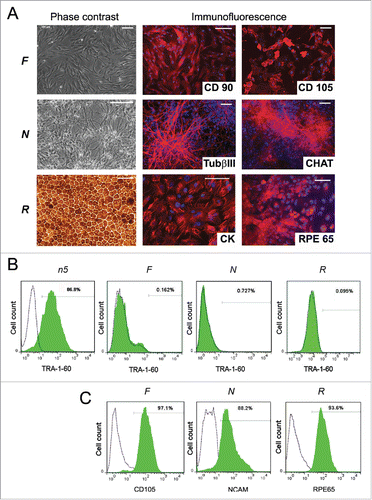
We established an isogenic system for reprogramming () by introducing reprogramming factors into the previously described hESC line hESM01.Citation18,19 The cell line hESM01-OSKMN-DOX-n5 (hereafter referred to as n5) that expressed all transgenes exclusively in the presence of DOX in undifferentiated or differentiated states, had a normal karyotype and demonstrated pluripotency in vitro and in vivo was selected for further analysis (Fig. S1 and Supplemental Experimental Procedures). The n5 cell line was used to generate 3 somatic cell lines: fibroblast-like cells (F), neuronal precursors (N), and retinal pigment epithelial cells (RPE, R). For the details of these procedures, see the Supplemental Experimental Procedures. To ensure that reprogramming would proceed only in differentiated somatic cells, magnetic separation was performed using antibodies against the markers CD31/CD105, NCAM, and RPE65 for the respective somatic cell populations. Specialized cell types were further analyzed for the presence of lineage-specific markers, the absence of PSC markers, and transgene induction (). To ensure that cell types differentiated from the n5 cell line closely resemble those chosen for the reprogramming, a somatic cell genome-wide transcriptome analysis was performed. Comparison of transcriptome data and available data sets confirmed that all 3 types of somatic cells expressed a set of cell type specific markers. The n5-derived fibroblasts closely resembled MRC5 (human lung fibroblasts), BJ1 (human foreskin fibroblasts) and human skin fibroblasts;Citation20,21 the n5-derived neurons corresponded to the human gray and white matter brain cells;Citation22 and the transcriptome of our RPE cells was similar to previously published hRPE cells.Citation23,24,25 (for details see Supplemental Experimental Procedures Fig. S2).
Figure 2. Characterization of n5 somatic derivatives. (A) Morphology (phase contrast microscopy) and immunocytochemistry of fibroblast-like cells (F), neurons (N), and retinal pigment epithelial cells (R). TUBb3, β-tubulin (III); CHAT, choline acetyltransferase; CK, cytokeratin; RPE65, retinal pigment epithelium-specific 65 kDa protein. Signals corresponding to the respective markers are shown in red, with blue indicating nuclei stained with DAPI. Scale bar, 100 mM. (B) FACS analysis of TRA-1-60 expression in differentiated cells. Dotted lines represent isotype controls and antibody staining is shown in green. (C) FACS analysis of the purity of magnetically separated F, N, and R cells for corresponding somatic markers.
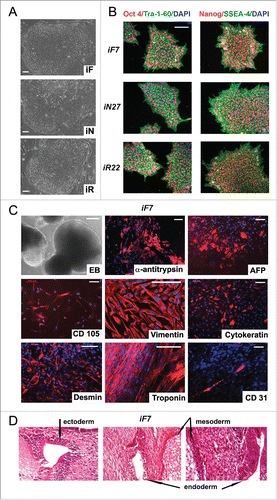
Differentiated cell lines were reprogrammed by adding DOX (). Importantly, all iPSC lines were generated using the same protocol in parallel. The average reprogramming efficacy in all 3 somatic cell types; i.e., the number of iPSC clones with respect to the number of cells in the starting population, was approximately 3%. Established iPSCs were further analyzed for pluripotency marker expression, somatic gene down regulation (Table S1), transgene silencing, karyotype, and in vitro and in vivo pluripotency (). Pairs of independently selected fibroblast-, neuron-, and RPE-derived iPSC lines (iF, iN, and iR, respectively) were used for further genome-wide analyses.
Figure 3. Characterization of isogenic iPSC lines reprogrammed from fibroblasts, neurons and RPE cells. (A) colony morphology of iF, iN, and iR cells; (B) immunocytochemical assay of iF, iN, and iR cells; for pluripotent markers (red and green indicate respective markers, blue indicates DAPI); (C) representative images of embryoid bodies and immunocytochemistry of in vitro iPSC-derived differentiated cell (red indicates markers, blue indicates DAPI) iF7 clones are shown. Scale bar, 100 mM; (D) Teratoma sections derived from the iF7 cell line, hematoxylin-eosin staining.
Genome-wide similarity of the global patterns of DNA methylation and gene expression in isogenic ESCs and iPSCs
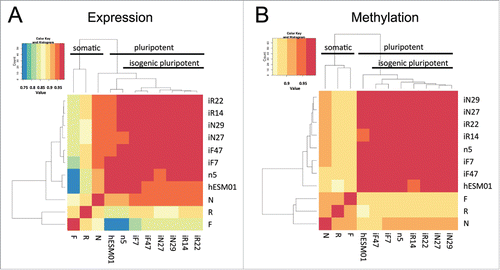
Genome-wide methods were used to perform a systematic comparison of DNA methylation (Infinium HumanMethylation450 BeadChip, Illumina) and gene transcription (HumanHT-12 v4 Expression BeadChip, Illumina) between 2 parental hESC lines (hESM01 and n5), 3 n5-derived somatic cell lines, and 3 pairs of iPSC clones (). All tested cell lines were isogenic according to the STR analysis (Table S2). We used previously developed tools and datasets to confirm that data generated from genome-wide analysis of the established isogenic cell lines separated them according to their biological properties (Fig. S4). A hierarchical clustering was performed to determine whether global patterns of DNA methylation and gene expression distinguish PSCs from ESС-derived somatic cells and divide iPSC lines into subclasses according to their somatic origin. Two distinct clusters emerged, one comprising all PSC lines (with a Spearman correlation of 95–98% within the PSC cluster) and the other comprising all somatic cell lines (with a Spearman correlation of 85–90% between somatic and PSC clusters) (, Table S2). Within the PSC cluster, iPSC lines derived from the same somatic cell type frequently clustered together (for example, 2 cell lines of fibroblast origin, iF7 and iF47). In our case, it could also be explained by the so-called one-dish batch effect, i.e., similarity based on culture in the same starting dish and the act of reprogramming, which could create a unique environment.
Figure 4. Genome-wide analysis of gene transcription and DNA methylation in the isogenic pluripotent and somatic cell lines. (A, B) Heatmaps based on Spearman correlations between all cell lines; the color scale ranges from 0.75 to 1. (A) Transcriptome and (B) methylome data (P < 0.01, FDR < 0.05).
Because our isogenic system employed multiple cell selection procedures (), we could not exclude the possibility that gene expression were altered simply by in vitro manipulations during these bottleneck procedures.Citation26 To identify genes that gradually increased or decreased their expression level during cell selection procedures, gene expression data from iPSC lines were compared with their parental somatic lines and the isogenic n5 ESC line. Only a few genes gradually increased or decreased transcription during technical manipulations, and none demonstrated altered transcriptional levels in all cell lines synchronously (data not shown).
Genome-wide analysis of the reprogramming process in different somatic lineages of the same origin
It was apparent that during pluripotent cell differentiation into somatic lineages and reversal of this state back to pluripotency (), significant changes occurred in the transcriptional and methylation profiles of reprogrammed somatic cells that were ultimately consolidated in a particular iPSC line. During this back-and-forth process, some genes and/or CpGs had similar changes in their expression and methylation profiles, revealing hallmarks of the process. We decided to combine genes and/or CpGs that changed their profiles synchronously in each independent iPSC type during the acquisition of pluripotency in 4 distinct groups (). Genes and/or CpGs that did not undergo a change in transcription and methylation levels in any cell type were considered intact. Genes and/or CpGs that maintained the same expression and/or methylation levels in established iPSC lines as in parental ESCs were designated as common for PSCs (CPSC). The somatic memory group was defined as a group in which genes and/or CpGs were expressed and methylated at the same level in iPSC lines and corresponding somatic cells (iF and F, iN and N, iR and R), but were different from that of ESCs. The clone-specific group comprised genes and/or CpGs that had an expression and/or methylation pattern in iPSC line distinct from the pattern observed for somatic cells and ESСs. CpG methylation and gene expression levels were considered independently (a difference in β value > 0.2 for DNA methylation or > 1.5-fold difference in expression level), and all data were assessed using a significance level of P < 0.01 and FDR < 0.05. Applying this grouping system independently to each iPSC pair derived from the same somatic cell type resulted in the list of genes and/or CpGs that have similar expression and methylation during their particular differentiation-reprogramming process (Table S3).
Figure 5. Reprogramming process, somatic memory and stochastic event input analysis in an isogenic system of human PSCs. (A) Graphical representation of the approach to analyze gene expression and CpG methylation during differentiation and reprogramming (“=” – same level of expression or methylation between indicated groups, “som” – somatic cell lines). (B) Input of genes (bold) and CpGs (italic) that drifted from specified groups (same colors as in A) into the “clone-specific” (red background) and “somatic memory” (blue background) groups during iPSC passaging. The number of passages is shown on top. (C) Clone-specific genes of different isogenic iPSC lines. (D) The methylation of 34 clone-specific CpGs is a specific reprogramming signature for our isogenic cell lines. The heatmap on the left shows that the methylation level of 34 CpG sites distinguishes iPSCs from ESCs and somatic cell lines in our isogenic system. However, the same CpGs do not discriminate ESCs and iPSCs in the independent GSE31848 data set; heatmap on the right. (E) Somatic memory group genes were identified for each iPSC type separately; however, most of them are shared by different iPSCs.
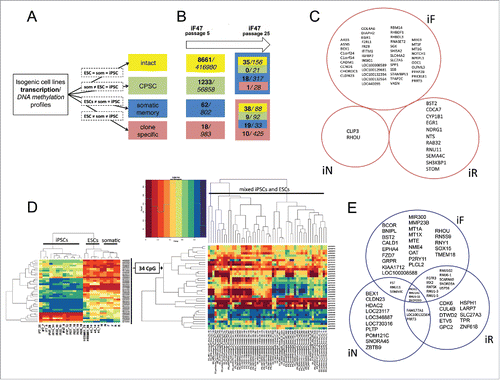
Surprisingly, most genes and/or CpGs showed no changes in expression and methylation during differentiation-reprogramming events in the analyzed cell lines (Fig. S5A). The expression of nearly 60% of the genes was unaltered, and most of the differentially expressed genes (>94%) belonged to the CPSC group. That is, their expression became ESC-like after reprogramming. Only 1–4% of differentially expressed genes belonged to the somatic memory group, with the same expression level in iPSC and parental somatic cell lines. CpG methylation was even more conservative during differentiation and reprogramming events; more than 85% of CpGs retained their original level of methylation. Most of the differentially methylated CpGs (> 95%) were also associated with the CPSC group, and only 1–2% of differentially methylated CpGs belonged to the somatic memory group. The number of clone-specific genes and/or CpGs was approximately the same as the number of somatic memory genes and/or CpGs; the maximum number of clone-specific CpGs was found in iR lines.
It was reported that iPSCs are not fully reprogrammed at early passages and additional passaging improves their properties.Citation27 Thus, it is obvious to speculate that somatic specific features (somatic memory + clone specific groups) of the cells will be reduced while the pluripotency-related signature (CPSC + intact groups) is unchanged or enriched. The established isogenic system enabled changes in particular genes and/or CpGs to be traced by determining from and to which group of specific genes and/or CpGs drifted during iPSC passaging. We decided to follow up changes that occurred during culturing and estimate their possible input in the iPCS molecular signature. Gene expression and DNA methylation patterns of fibroblast-derived iPSC lines from early (4–12) and late (25–29) passages were examined (, Table S4). Surprisingly, we found that the number of genes in the somatic memory group remained the same over multiple passages, although the number of clone-specific genes increased from 18 to 76. At the same time, the number of CpGs in the somatic memory and clone-specific groups decreased by approximately one-third. In the clone-specific and somatic memory groups, methylation was unchanged in 67% and 61% of CpGs, respectively, whereas only 13% of clone-specific and 33% of somatic memory genes remained fixed in these groups. The major contributor to the reprogrammed cell signature came from genes or CpGs (approximately 60% and 30%, respectively) that drifted from the early passage intact and CPSC groups, which both reflect ESC-like properties. This finding demonstrates that the establishment of pluripotency balance at early passages is accompanied by bidirectional changes in gene expression and/or CpG methylation. Only a small fraction of clone-specific and somatic memory genes and/or CpGs (less than one-third) that distinguish iPSCs from ESCs acquired and maintained their profiles during reprogramming and pluripotency establishment; the others are likely the result of stochastic fluctuations with no apparent biological significance.
To identify the reprogramming-specific signature in more detail, we compared the expression and methylation levels in pairs of iPSC lines to their corresponding somatic predecessor and maternal ESCs (). We found a small set of genes and/or CpGs for which the expression and methylation level was specific for each iPSC type (i.e., clone-specific group) (Table S3). To examine the functional significance of individual CpGs located in close proximity to gene promoters, we combined CpGs in CpG loci on the basis of their location, and found CpG loci belonging to the clone-specific group of each iPSC type (iFs, iNs, and iRs). It is worth noting that both the expression and methylation patterns of one of the best candidate reprogramming-specific genes, MEG3,Citation8,27,28 were unique in iFs but were similar to ESCs in iNs and iRs, confirming its inconsistent role as a universal marker of reprogramming. To determine the reprogramming specific signature, we assumed that genes and/or CpG + CpG loci belonging to a clone-specific group of each iPSC type will contain such marks. We did not find any genes that were common to a clone-specific group of all types of iPSCs (), although 34 CpGs and one CpG locus (the CpG island of the CBLN4 gene) had a unique methylation pattern in all our iPSC lines. To verify whether this CpG signature is also characteristic for other human PSC lines, these CpGs and CpG loci were used to distinguish between ESCs and iPSCs in the GSE31848 data set.Citation16 There were no clear ESC and iPSC clusters or altered methylation of the CBLN4 gene in iPSCs among pluripotent cells (). Interestingly, the CBLN4 gene was the most frequently demethylated in 122 hESC lines analyzed at early and late passages in various laboratories,Citation19 which indicates a high heterogeneity in its methylation level among even “gold standard” pluripotent lines. Taken together, these data demonstrate that the observed reprogramming-specific signature in iPSCs mostly results from fluctuations that are likely to be introduced by the laboratory-specific environment and not by the reprogramming process itself.
Isogenic iPSCs do not have somatic specific memory and possess a “core” pluripotency signature
The expression and methylation data clearly distinguished iPSCs from ESCs (). This implies that each iPSC line differs from the others not only by a small clone-specific set of genes and/or CpGs but also by their differentiated origin or even by the CPSC gene and/or CpG pattern. Therefore, we asked whether successful reprogramming always generates cells with the same molecular pluripotency status or the starting cell type, and whether other factors can lead to differences between functionally similar pluripotent stem cells.
The somatic memory group of genes and/or CpGs was previously defined as being specific for iFs, iNs, iRs, and the corresponding somatic cells, however different from the parental ESC line. In fact, these differences may affect the potential usefulness of iPSCs and may even present a disadvantage in some cases. In the isogenic system, a small number of genes (up to 40 in iFs) and CpGs (up to 927 in iRs) belonged to this group (). Moreover, within the somatic memory group, 20 genes and 338 CpGs were shared by at least 2 of 3 iPSC types and therefore could not be considered as reflecting a particular cell type of origin. This fact provides additional support for the hypothesis that there is a set of genes and/or CpGs that reflects the memory of a general differentiated state.Citation7 Additionally, we tested whether CpGs found in the somatic memory group in our iFs contained CpGs specific for fibroblast-derived iPSCs that were recently published.Citation29 Only 3 CpGs were common in the 2 datasets, demonstrating the impact of the laboratory or general cell line variations to the final iPSC DNA methylation status. To determine whether somatic memory genes and/or CpGs unique to each iPSC type reflected the specific cell type, a Gene Ontology analysis of these genes and/or CpGs was carried out. We did not find an enrichment of functions or processes specific to a particular cell type; in addition, a manual investigation of somatic memory genes did not reveal any indications on specific to somatic cell type genes. We therefore conclude that iPSC does not have a somatic cell type specific memory; however, it does carry unspecific signatures (genes and/or CpGs) reflecting a preexisting differentiated state or ESCs heterogeneity. Molecular differences in the pluripotent status of each iPSC type were evaluated by analyzing sets of genes and/or CpGs belonging to the CPSC group of iFs, iRs, and iNs. Unexpectedly, only a limited number of genes and/or CpGs was shared by all iPSC lines, which were designated as the core set (Table S5, Fig. S5C). To determine the biological significance of this set of genes and/or CpGs as well as those shared by pairs of iPSCs or belonging to a single iPSC type, the GREAT, GOrilla, and WebGestalt tools were employed. The enrichment data is shown in Table S6.
The core set was enriched for genes involved in the regulation of epithelial cell proliferation. Likewise, CpGs in the core set were associated with epithelial differentiation and neuronal commitment, including hypomethylation of Pax6 and Pax3, the main transcription factors in neural crest development. In addition, we also observed enrichment for targets of the ESC-specific epigenetic regulators H3K27me3 and Polycomb, as well as targets of transcription factors that regulate pluripotency, such as Oct4, Nanog, and Sox2.
We analyzed the functions of CPSC genes shared by any 2 types of iPSCs and determined that cell cycle, proliferation, and mitotic processes were enriched (). The remaining CPSC genes and/or CpGs unique to each iPSC type showed enrichment in metabolic processes (iF and iR) and the immune response (iR and iN). We did not detect enrichment for targets of ESC-associated transcription factors and regulators among iN- and iR-specific CPSC genes, indicating that they are not directly involved in the major ESC-specific functions. Moreover, these CPSC genes and/or CpGs have ESC-specific patterns of expression/methylation that coordinate the self-renewal of newly generated iPSCs. Given the diversity of ESC-like genes and/or CpGs, this result demonstrates that in each reprogramming event, a unique set of changes leads to the acquisition of an ESC-like state.
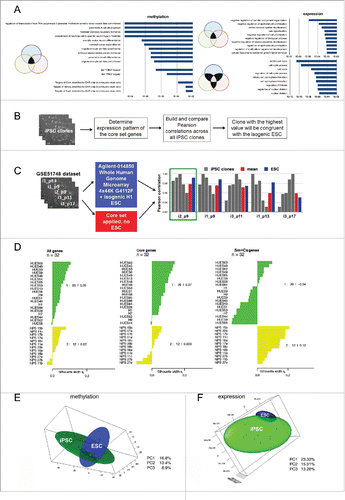
Figure 6. The isogenic system provides a core genetic signature for reprogrammed cells. (A) Top 10 Gene Ontology Biological Processes and Transcription Factor Targets shared by at least 2 different iPSC lines; the corresponding p-values are shown in bars. (B) Suggested approach to select an iPSC clone identical to its isogenic ESCs using the core gene set expression profile. (C) Examination of the best clone selection approach using the GSE51748 dataset. Pearson correlations between a particular iPSC clone and other clones calculated on the basis of the core set gene expression levels are shown in gray; the mean correlation between them is shown in red; Pearson correlations between particular iPSC clones and isogenic ESC lines calculated on the basis of all transcriptome data are shown in blue. The green rectangle indicates the best clone that has the highest mean correlation with others using both approaches. (D) Silhouette values (Sv) indicate the similarity of 32 ESCs (green cluster) and iPSCs (yellow cluster) lines from GSE25970 data set. On the right side, the Svs for each cluster are shown. Sv values were calculated using the expression pattern of all genes in the GSE25970 dataset (“All genes”): expression level only of the core set of genes; (“Core set”): expression level of the genes belonging to somatic memory and clone specific groups (“Sm+Cs genes”). (E, F) PCA analysis of isogenic PSC lines established in our study based on (E) methylation and (F) expression data. In both cases, 3 PCs were used and 3-D maps were built. The green sphere shows the location of the iPSC cluster, while the blue sphere shows the ESC cluster.
Whole-genome expression data predicts the minimal number of iPSC lines for analysis while a defined set of genes indicates their virtual ESC similarity
We did not find any significant input of somatic cell types into the isogenic iPSC lines that we studied, although the specific core set of genes and/or CpGs reflecting the pluripotent nature of iPSCs was defined. These findings prompted the question of how universal the gene set is and whether it can predict the similarity between any human iPSC line with its real or virtual isogenic ESCs (). We used this core set of genes to investigate whether this set could be applied to characterize the ESС-like properties of iPSC lines generated in other laboratories.
Recently, the GSE51748 data set consisting of microarray data from iPSC lines independently generated from neural progenitor cells by lentiviral transduction and the parental partly isogenic human ESC line from which the neural progenitors were differentiated were published.Citation30 We decided to apply a core set genes to predict which of the newly derived neural iPSCs was more similar to the parental ESC line and would therefore presumably be ideal for further applications. All newly derived iPSC clones passed the pluripotency tests (pluripotency marker expression, karyotyping, and in vivo and in vitro differentiation); therefore, they were presumed to be more or less similar to the parental ESC line and consequently acquired an isogenic ESC pattern of expression of our core set of genes. Since all human iPSC lines tend toward more isogenic ESC-like patterns of expression, it was hypothesized that the one that was most consistent with other iPSC clones in terms of expression of the core set of genes would be closer to their own real isogenic ESC line. We estimated Pearson correlations between iPSC clones in the GSE51748 dataset on the basis of the expression level of the core set of genes (Table S5, ). The iPSC clone NPC-i2 from the GSE51748 data set was the most consistent (i.e., had the highest mean correlation between all iPSC clones). The same clone had the highest correlation with the partly isogenic hESC line using whole-genome data from the GSE51748 dataset (). Notably, for almost all given cell lines, a higher correlation between iPSC lines indicates a higher correlation between this line and parental ESCs (with a Pearson's product-moment correlation of 0.72, Table S5). This result indicates the accuracy of the chosen method of prediction. A test of an independent data set demonstrates that we can effectively use a deduced core set of genetic markers to predict which particular iPSC line is closest to its theoretical isogenic ESCs.
Next, we decided to test the versatility of our core set for allogenic PSCs. In this case, we did not want to identify the iPSC clone most similar to some allogenic ESCs, but to measure whether our core set distinguishes the iPSCs subgroup with a similar core set expression pattern that is more similar to ESCs. To measure this we used the “silhouette” component analysis, where the average distance is calculated for every point of a cluster to all other points of the same cluster. A principal component analysis (PCA) was carried out using our transcriptome expression profiles of fibroblast-derived iPSCs and a variety of ESCs with different genotypes (GSE25970; Citation4). The cluster specific silhouette value (Sv) for iPSC lines between −1 and 1 measures how tightly a particular cluster is grouped and how well it is separated, which indicates how appropriately the data are clustered: the higher the number, the better the cluster is distinguished from the others. PCA was performed for all PSC lines from the GSE25970 dataset using the whole-genome expression profile, the “core set,” and a pooled set of genes from the “clone-specific” and “somatic memory” groups from our data set (Fig. S6). When we applied whole-genome data for the PCA, the Sv for iPSC cluster was 0.022, indicating that the reprogrammed cells were very close to the ESC lines. Nonetheless, using the “core set” genes, the iPSC cluster became nearly indistinguishable from the ESC cluster at Sv = 0.003. As a negative control for our quality prediction approach, we decided to apply a pooled set of genes from the “clone-specific” and “somatic memory” groups from our dataset. In this case, an iPSC cluster far distinct from ESC cells with Sv = 0.12 was formed (). Thus, iPSC lines from the independent GSE25970 data set also possessed the genetic signatures deduced in our study. Summarizing our data, we can conclude that we identified a set of genes that could be used to identify reprogrammed somatic cell lines most similar to their virtual parental ESC line. Notably, the identification is irrespective of the expression detection method and iPSCs somatic origin, which means that one could apply our “core set” genes to any iPSC lines from the same cohort to find a best clone in terms of similarity with the virtual parental ESC line.
We also estimated the minimum number of human iPSC clones that should be analyzed to obtain with 95% confidence at least a single cell line that perfectly matches its virtual ESCs. The PCA approach was used to reduce the dimensionality of the data and to globally visualize data from transcription profiling. A projection of the expression pattern onto the PCs separates individual cell lines into 2 distinct clusters of ESC and iPSC lines (). The shapes of these 3-D spheres represent variability between individual cell lines for pluripotent cell types. Cell lines that fall into the region of overlap between the 2 spheres (95% confidence interval, 2.7 σ) were indistinguishable based on their transcriptional profiles, and therefore, iPSC lines in this region cannot be discriminated from an ESC line. Such intersections are typically only observed for large datasets for which at least several dozen samples are analyzed; Citation4 however, the isogenic system allowed the input number of cell lines to be minimized. Using our data set, and in particular the data pertaining to inter-clone variability of iPSC lines and their distance to isogenic ESCs, we calculated that 5 randomly selected iPSC clones are sufficient to establish overlapping with ESCs; that is, among these 5 clones, at least one (+/− 2.7 σ) would be indistinguishable from isogenic ESCs with 95% confidence. Thus, 5 independently selected iPSC clones comprise the minimum number of cell lines that are required to analyze the similarity between iPSCs and ESCs. Additionally, we have identified a core set of genes whose expression levels could be used to identify the best iPSC clone in the cohort.
Discussion
One of the most important questions regarding the reprogramming of somatic cells to pluripotency is whether human iPSCs differ from ESCs in their properties and potential. It is clear that currently used criteria (immunological markers, pluripotency gene expression, DNA methylation level, teratoma formation) make iPSC lines indistinguishable from ESC lines. However, the need to differentiate a particular iPSC line into multiple lineages raises the issue of iPSCs quality in respect to their genotype-specific pluripotent state. Recently, a clinical trial utilizing iPSC-derived RPE cells was initiated. Twenty-four lines were screened to choose the most patient compatible cells.Citation31 It is now evident that differences in the quality of iPSC clones are largely due to technical variables relating to reprogramming approaches and culture conditions.Citation4,8,27,32 Additionally, some uncontrolled stochastic events during reprogramming undoubtedly influence gene expression and DNA methylation patterns in even functionally identical iPSC lines. Therefore, the evaluation of parameters that make iPSC line(s) indistinguishable from currently virtual but pre-existing ESCs, as well as the selection process, will be essential for identifying iPSC clones that are suitable for medical applications. To evaluate these parameters and to assess the influence of the technical aspects of reprogramming, we developed a complete isogenic system of human ESCs, along with their differentiated somatic derivatives and reprogrammed cell lines. Sensitive genome-wide analytical approaches demonstrated that even double bottleneck selection (cell selection upon differentiation and iPSC clones pick-up) did not introduce stepwise heritable changes affecting cell state.
Interestingly, we did not identify any genes that were common to all isogenic iPSC types and could distinguish them from the isogenic ESCs. The small number of CpGs shared by cell lines was rather laboratory-specific and did not represent a common trace of the reprogramming process for the isogenic iPSCs. Thus, there were no traces of reprogramming common to all iPSCs that can distinguish them from ESCs in a single set of isogenic lines they likely do not exist for other lines. However, differences between iPSC and ESCs in gene expression do exist (Table S4) and are likely to be universal for any other cell lines. Using this set of genes, we observed better segregation between the iPSC and ESC clusters in the GSE25970 dataset. This set comprises genes with well-known effects on the reprogramming process, such as Meg3 and Notch1.Citation33,34 Additionally, it contains 5 genes from the metallothionein family, all of them located within a 50 kb region on chromosome 16, therefore indicating an involvement of this loci in the reprogramming process. Taken together, our data suggest that in iPSCs, mostly stochastic expression of the genes that are rarely found in ESCs is observed, although some genes aberrantly expressed in iPSCs could be effectively used to qualify reprogrammed cells.
Recent advances in iPSCs application in disease treatment and discovery of the alternative stem cell-like states during reprogrammingCitation31,35 support the need for efficient and informative approaches to the selection of the best iPSC line in a cohort of functionally similar reprogrammed cells. Screening of dozens of clones that passed the teratoma assay by in vitro differentiation into a required somatic cell line to find the perfect clone for a specific application has not been effective Citation36,37 and considering the possible need for multiple somatic cell types. Even the previously developed scorecard approach may be inefficient in the search for iPSC clone that would match with a patient's own isogenic ESC line. Differentiation into a variety of cell types is the intrinsic property of ESCs; therefore, choosing the iPSC line most identical to its preexisting ESCs is the way to identify a universal iPSC clone suitable for differentiation in multiple directions. In our study, we identified a core set of genes whose expression level justifies similarity of reprogrammed somatic cells to ESC not only in our isogenic system but also for iPSCs generated in any independent experiments. Finally, we calculated a minimum number of iPSC clones required for the similarity analysis. At least 5 independent clones have to be established, analyzed functionally, and tested using our core gene set to identify the clone that would match their (theoretical) isogenic hESCs. Summarizing our findings, we can conclude that human iPSCs and ESCs are very similar, although each act of reprogramming leads to the acquisition of a pluripotent state specific for each iPSC and a rather small number of genetic markers can be utilized to predict those most similar to the ESC state.
Materials and methods
ESCs isogenic system establishment and characterization
The hESM01 cells were transduced with lentiviruses containing genes for 5 transcription factors (Oct4, Sox2, KLF4, c-Myc, and Nanog, under the control of the DOX- inducible promoter and neomycin resistance). The cells were selected for G418, cloned and analyzed for all 5 transcription factor insertions, their induction upon DOX addition and silencing upon withdrawal in undifferentiated and differentiated states. ESC clones that met these conditions were analyzed for genome integrity and pluripotency maintenance in vitro and in vivo (see Supplemental Experimental Procedures).
Differentiation of human ESCs
Differentiation of the n5 cell line into fibroblast-like cells, RPE cells and neural cells, magnetic selection, FACS and genome-wide transcriptome analyses are described in detail in the Supplemental Experimental Procedures.
Reprogramming
On the first day of reprogramming, the medium for all 3 types of differentiated cells was changed to an ESC medium with the addition of 1 mg/ml doxycycline (Stemgent). On post-induction day 8-12, the first clones appeared. On day 18-25, ESC-like colonies were picked up on a Matrigel-coated 24-well plate in a doxycycline-free mTeSR1 medium.
Methylation and expression data profiling
Two ESC lines, 3 somatic cell lines, pairs of iN and iR iPSC lines, and 2 iF lines from different passages (“early” and “late”) were analyzed using Infinium 450K BeadChips and HT-12v4 Expression BeadChips (both from Illumina, Inc.). Manufacturer protocols for probe preparation and processing were used. In GenomeStudio, the probes were quality controlled and filtered for those detected at p < 0.01 in at least one sample and exported for normalization in R (cran.r-project.org). ComBat was used for batch effect elimination Citation38 on both data sets. A peak correction of the 450k dataset was performed using the pipeline from the research of Touleimat and Tost.Citation39 The IMA, limma, and lumi R packages were used to analyze differential expression (2-sample t-test) or methylation (Mann-Whitney test) between groups of samples. In both cases, p-values < 0.01 and a Benjamini-Hochberg false discovery rate corrected q-value < 0.05 were used. Expression changes more than 2-fold and β-value differences more than 0.2 were considered significant. The gene ontology term analysis was performed using GREAT,Citation40 Gorilla,Citation41 and WebGestalt Citation42 tools. 3D principal component analyses (ellipsoids) of the expression data were constructed on the basis of the covariance matrix with 2 standard deviation scaling. All data are available by the reference series GSE70739 from GEO repository.
Calculation of minimal number of iPSC lines for the analysis
The minimal number of iPSC clones was calculated as follows: using the normality of the given PC1 distribution for ESCs and iPSCs, the mean and sigma for each case were found. By integrating the formula of normal distribution for iPSCs average and variance, the probability of hitting iPSCs in the ESC zone was calculated. The integration was performed on a plot mean +/− 2.7 * sigma. On the basis of this probability using a binomial distribution, the minimal number of iPSC clones that would be enough for at least one hitting the ESC zone with 95% confidence was calculated.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary Files
Download MS Word (6.4 MB)Acknowledgments
We acknowledge Dr. Hochedlinger K. and his laboratory where Addgene plasmids were constructed. We are grateful to Dr. A. Tomilin for his help with teratoma assays, Dr. E. Philonenko for critical review of the manuscript. We acknowledge JetBrains Inc. and particularly O. Shpinev and colleagues as well as A. Panchin from The Institute for Information Transmission Problems RAS for their help with methylation data analysis.
Funding
This study was supported by FASO intramural research funding IV-53.10, IV-53.37 and FRBMT. Experiments with RPE cells were supported by Russian Scientific Foundation Grant # 14-15-00930
References
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science 1998; 282:1145-7; PMID:9804556; http://dx.doi.org/10.1126/science.282.5391.1145
- Pan CK, Heilweil G, Lanza R, Schwartz SD. Embryonic stem cells as a treatment for macular degeneration. Expert Opin Biol Ther 2013; 13:1125-33; PMID:23705996; http://dx.doi.org/10.1517/14712598.2013.793304
- Schwartz SD, Hubschman J-P, Heilwell G, Franco-Cardenas V, Pan CK, Ostrick RM, Mickunas E, Gay R, Klimanskaya I, Lanza R. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet 2012; 379:713-20; PMID:22281388; http://dx.doi.org/10.1016/S0140-6736(12)60028-2
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 2011; 144:439-52; PMID:21295703; http://dx.doi.org/10.1016/j.cell.2010.12.032
- Marchetto MCN, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One 2009; 4:e7076; PMID:19763270; http://dx.doi.org/10.1371/journal.pone.0007076
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 2009; 5:111-23; PMID:19570518; http://dx.doi.org/10.1016/j.stem.2009.06.008
- Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol 2011; 13:541-9; PMID:21499256; http://dx.doi.org/10.1038/ncb2239
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 2010; 465:175-81; PMID:20418860; http://dx.doi.org/10.1038/nature09017
- Nostro MC, Keller G. Generation of beta cells from human pluripotent stem cells: Potential for regenerative medicine. Semin Cell Dev Biol 2012; 23:701-10; PMID:22750147; http://dx.doi.org/10.1016/j.semcdb.2012.06.010
- Atala A, Kasper FK, Mikos AG. Engineering complex tissues. Sci Transl Med 2012; 4:160rv12; http://dx.doi.org/10.1126/scitranslmed.3004890
- Tedesco FS, Gerli MFM, Perani L, Benedetti S, Ungaro F, Cassano M, Antonini S, Tagliafico E, Artusi V, Longa E, et al. Transplantation of genetically corrected human iPSC-derived progenitors in mice with limb-girdle muscular dystrophy. Sci Transl Med 2012; 4:140ra89; PMID:22745439; http://dx.doi.org/10.1126/scitranslmed.3003541
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol 2010; 28:848-55; PMID:20644536; http://dx.doi.org/10.1038/nbt.1667
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 2011; 470:68-73; http://dx.doi.org/10.1038/nature09798
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LIR, et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010; 467:285-90; PMID:20644535; http://dx.doi.org/10.1038/nature09342
- Ruiz S, Panopoulos AD, Montserrat N, Multon MC, Daury A, Rocher C, Spanakis E, Batchelder EM, Orsini C, Deleuze JF, et al. Generation of a drug-inducible reporter system to study cell reprogramming in human cells. J Biol Chem 2012; 287:40767-78; PMID:23019325; http://dx.doi.org/10.1074/jbc.M112.384024
- Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Müller F-J, Wang Y-C, Boscolo FS, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell 2012; 10:620-34; PMID:22560082; http://dx.doi.org/10.1016/j.stem.2012.02.013
- Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell 2012; 10:678-84; PMID:22704507; http://dx.doi.org/10.1016/j.stem.2012.05.005
- Lagarkova MA, Volchkov PY, Lyakisheva AV, Philonenko ES, Kiselev SL. Diverse epigenetic profile of novel human embryonic stem cell lines. Cell Cycle 2006; 5:416-20; PMID:16479162; http://dx.doi.org/10.4161/cc.5.4.2440
- International Stem Cell Initiative, Amps K, Andrews PW, Anyfantis G, Armstrong L, Avery S, Baharvand H, Baker J, Baker D, Munoz MB, et al. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol 2011; 29:1132-44; PMID:22119741; http://dx.doi.org/10.1038/nbt.2051
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 2010;42:1113-7; PMID:21057500; http://dx.doi.org/10.1038/ng.710
- Wang XM, Yik WY, Zhang P, Lu W, Dranchak PK, Shibata D, Steinberg SJ, Hacia JG. The gene expression profiles of induced pluripotent stem cells from individuals with childhood cerebral adrenoleukodystrophy are consistent with proposed mechanisms of pathogenesis. Stem Cell Res Ther 2012; 3:39; PMID:23036268; http://dx.doi.org/10.1186/scrt130
- Murrell W, Palmero E, Bianco J, Stangeland B, Joel M, Paulson L, Thiede B, Grieg Z, Ramsnes I, Skjellegrind HK, et al. Expansion of multipotent stem cells from the adult human brain. PLoS One 2013; 8:e71334; PMID:23967194; http://dx.doi.org/10.1371/journal.pone.0071334
- Zhang Z, Zhang Y, Xiao H, Liang X, Sun D, Peng S. A gene expression profile of the developing human retinal pigment epithelium. Molecular Vision 2012; 18:2961-75; PMID:23487591
- Strunnikova NV, Maminishkis A, Barb JJ, Wang F, Zhi C, Sergeev Y, Chen W, Edwards AO, Stambolian D, Abecasis G, et al. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum Mol Genet 2010; 19:2468-86; PMID:20360305; http://dx.doi.org/10.1093/hmg/ddq129
- Booij JC, van Soest S, Swagemakers SM, Essing AH, Verkerk AJ, van der Spek PJ, Gorgels TG, Bergen AA. Functional annotation of the human retinal pigment epithelium transcriptome. BMC Genomics 2009; 10:164-178; PMID:19379482; http://dx.doi.org/10.1186/1471-2164-10-164
- Bhatia S, Pilquil C, Roth-Albin I, Draper JS. Demarcation of stable subpopulations within the pluripotent hESC compartment. PLoS One 2013; 8:e57276; PMID:23437358; http://dx.doi.org/10.1371/journal.pone.0057276
- Carey BW, Markoulaki S, Hanna JH, Faddah DA, Buganim Y, Kim J, Ganz K, Steine EJ, Cassady JP, Creyghton MP, et al. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell 2011; 9:588-98; PMID:22136932; http://dx.doi.org/10.1016/j.stem.2011.11.003
- Stelzer Y, Ronen D, Bock C, Boyle P, Meissner A, Benvenisty N. Identification of novel imprinted differentially methylated regions by global analysis of human-parthenogenetic-induced pluripotent stem cells. Stem Cell Reports 2013; 1:79-89; PMID:24052944; http://dx.doi.org/10.1016/j.stemcr.2013.03.005
- Huang K, Shen Y, Xue Z, Bibikova M, April C, Liu Z, Cheng L, Nagy A, Pellegrini M, Fan J-B, et al. A Panel of CpG Methylation Sites Distinguishes Human Embryonic Stem Cells and Induced Pluripotent Stem Cells. Stem Cell Reports 2014; 2:36-43; PMID:24511466; http://dx.doi.org/10.1016/j.stemcr.2013.11.003
- Mallon BS, Hamilton RS, Kozhich OA, Johnson KR, Fann YC, Rao MS, Robey PG. Comparison of the molecular profiles of human embryonic and induced pluripotent stem cells of isogenic origin. Stem Cell Res 2014; 12:376-86; PMID:24374290; http://dx.doi.org/10.1016/j.scr.2013.11.010
- Assawachananont J, Mandai M, Okamoto S, Yamada C, Eiraku M, Yonemura S, Sasai Y, Takahashi M. Transplantation of embryonic and induced pluripotent stem cell-derived 3D retinal sheets into retinal degenerative mice. Stem Cell Reports 2014; 2:662-74; PMID:24936453; http://dx.doi.org/10.1016/j.stemcr.2014.03.011
- Rouhani F, Kumasaka N, de Brito MC, Bradley A, Vallier L, Gaffney D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet 2014; 10:e1004432; PMID:24901476; http://dx.doi.org/10.1371/journal.pgen.1004432
- Stadtfeld M, Apostolou E, Ferrari F, Choi J, Walsh RM, Chen T, Ooi SSK, Kim SY, Bestor TH, Shioda T, et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat Genet 2012; 44:398-405; PMID:22387999; http://dx.doi.org/10.1038/ng.1110
- Ichida JK, TCW J, Williams LA, Carter AC, Shi Y, Moura MT, Ziller M, Singh S, Amabile G, Bock C, et al. Notch inhibition allows oncogene-independent generation of iPS cells. Nat Chem Biol 2014; 10:632-9; PMID:24952596; http://dx.doi.org/10.1038/nchembio.1552
- Tonge PD, Corso AJ, Monetti C, Hussein SMI, Puri MC, Michael IP, Li M, Lee D-S, Mar JC, Cloonan N, et al. Divergent reprogramming routes lead to alternative stem-cell states. Nature 2014; 516:192-7; PMID:25503232; http://dx.doi.org/10.1038/nature14047
- Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol 2011; 29:279-86; PMID:21293464; http://dx.doi.org/10.1038/nbt.1783
- Daley GQ, Lensch MW, Jaenisch R, Meissner A, Plath K, Yamanaka S. Broader implications of defining standards for the pluripotency of iPSCs. Cell Stem Cell 2009; 4:200-1; PMID:19265657; http://dx.doi.org/10.1016/j.stem.2009.02.009
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2006; 8:118-27; PMID:16632515; http://dx.doi.org/10.1093/biostatistics/kxj037
- Touleimat N, Tost J. Complete pipeline for Infinium(→) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics 2012; 4:325-41; PMID:22690668; http://dx.doi.org/10.2217/epi.12.21
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010; 28:495-501; PMID:20436461; http://dx.doi.org/10.1038/nbt.1630
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 2009; 10:48; PMID:19192299; http://dx.doi.org/10.1186/1471-2105-10-48
- Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 2013; 41:W77-83; PMID:23703215; http://dx.doi.org/10.1093/nar/gkt439

