?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Centrosomal proteins play important roles in the spindle assembly and the segregation of chromosomes in the eukaryotic cells. STARD9, a recently identified centrosomal protein, was reported to influence the spindle pole assembly. However, the role of STARD9 in maintaining the stability and organization of microtubules are not known. Here, we show that STARD9 regulates the assembly and dynamics of both interphase and mitotic microtubules. The knockdown of STARD9 in HeLa or HCT116 cells with siRNA or shRNA induced a strong depolymerization of the interphase microtubules. The over-expression of the motor domain of STARD9 stabilizes microtubules against cold and nocodazole suggesting that STARD9 stabilizes microtubules in HeLa cells. Using fluorescent recovery after photobleaching, we showed that the knockdown of STARD9 strongly reduced microtubule dynamics in the live spindles of HeLa cells. The reassembly of microtubules in the STARD9-depleted cells was strongly reduced as compared to the microtubules in the control cells implying the role of STARD9 in the nucleation of microtubules. Further, the depletion of STARD9 inhibited chromosome separation and the STARD9-depleted HeLa cells were blocked at mitosis. Interestingly, the frequency of multipolar spindle formation increased significantly in the STARD9-depleted HeLa cells in the presence of vinblastine and the STARD9-depleted cells showed much higher sensitivity towards vinblastine than the control cells indicating a new approach for cancer chemotherapy. The evidence suggests that STARD9 regulates the assembly and stability of both interphase and spindle microtubules and thereby, play important roles in the cell cycle progression.
Introduction
Several centrosomal proteins have been found to have important roles in the cell cycle progression [Citation1–Citation7]. STARD9, (steroidogenic acute regulatory protein-related lipid transfer (START) domain containing 9), is one of the newly identified centrosomal proteins that binds to mitotic microtubules and thought to have a role in spindle pole assembly [Citation8,Citation9]. STARD9 is present in most of the tissues such as skin, brain, and heart. STARD9 depletion was reported to induce multipolar spindle formation due to the fragmentation of the pericentriolar matrix [Citation9]. Interestingly, the multipolar cells formed upon STARD9 depletion in case of cancer cells were significantly higher than the depletion effect in normal cells. However, no direct correlation was observed between the expression level of the protein and the formation of multipolar spindles [Citation9]. The STARD9-depleted HeLa cells were reported to activate the spindle assembly checkpoint proteins by increasing the localization of BubR1 and Bub1 at the kinetochores [Citation9]. The components of the chromosomal complex (AurKB, INCENP) required for mitotic arrest was also found to be present at the kinetochores indicating that the depletion of STARD9 activates the spindle assembly checkpoint [Citation9]. STARD9 consists of a motor domain, a forkhead-associated domain, a coil-coiled domain and the START domain. The N-terminal motor domain of STARD9 (STARD9-MD) is reported to bind to microtubules and to show ATPase activity [Citation9,Citation10]. The expression of STARD9-MD in HeLa cells induced pericentriolar matrix (PCM) fragmentation, suggesting a role of the motor domain in the spindle assembly [Citation9]. Microtubule-mediated forces are considered to be responsible for maintaining the cohesive force at the PCM, which if altered results in multipolar spindle formation due to the fragmentation of PCM [Citation2,Citation6,Citation11,Citation12]. We hypothesize a role for STARD9 in regulating microtubule assembly and another role in providing the balanced force for proper segregation of the chromosomes.
Earlier, the N-terminal motor domain of STARD9 was shown to bind to microtubules and taxol and nocodazole were found to reduce the effect of the depletion of STARD9 on PCM fragmentation in HeLa cells [Citation9]. However, the role of STARD9 on the organization and stability of interphase microtubules was not examined in the previous study [Citation9]. Further, the role of STARD9 in the regulation of the dynamics of spindle microtubules is not known. In this study, we report that the depletion of STARD9 induced microtubule depolymerization in HeLa and HCT116 cells. In addition, the expression of the N-terminal motor domain of STARD9 (STARD9-MD) in HeLa cells stabilized microtubules against both cold and nocodazole-induced disassembly. However, the expression of a mutant (R223A) STARD9-MD construct failed to stabilize the microtubules. Further, the spindle microtubule dynamics was found to be strongly suppressed in the STARD9-depleted HeLa cells. The chromosome separation was also strongly inhibited in the STARD9-depleted mitotic HeLa cells. In addition, STARD9-depleted HeLa cells were much more sensitive to low concentration of vinblastine than the control cells indicating that the depletion of STARD9 can be combined with an inhibitor of microtubules as an attractive strategy for the treatment of cancer.
Results
STARD9 is associated with tubulin polymeric fraction of HeLa cell extract
Cyclin B1 was found in the mitotic cell extract while it was not detectable in the interphase extract (Supplementary Figure1A). STARD9 was found to be present in similar level both in interphase and mitotic HeLa cells extract (Supplementary Figure1A). Both polymeric and soluble fractions of tubulin extracts of HeLa cells contained STARD9 (Supplementary Figure1B). Interestingly, the level of STARD9 was found to be 40 ± 9% higher in the polymeric tubulin fraction than the soluble tubulin fraction (Supplementary Figure1B). To check the binding of STARD9 to tubulin, we performed an immunoprecipitation assay using tubulin IgG. Tubulin IgG was found to pull down STARD9 from the polymeric tubulin fraction while GAPDH IgG failed to pull down STARD9 indicating that STARD9 binds to microtubules (Supplementary Figure1C). Under similar conditions, GAPDH IgG pulled down GAPDH from the cell extract indicating the specificity of the reaction (Supplementary Figure1C).
STARD9-MD binds to microtubules
Co-immunostaining experiment indicated that STARD9-MD co-localizes along the length of interphase microtubules while a STARD9-MD mutant (R223A) could not co-localize on microtubules (Supplementary Figure2A). The GFP transfected HeLa cells also did not show any co-localization on microtubules (Supplementary Figure2A). In addition, tubulin IgG was found to pull down GFP-STARD9-MD from the cell extract while GAPDH IgG failed to pull down GFP-STARD9-MD (Supplementary Figure2B). Previously, STARD9-MD was reported to bind to microtubules in vitro while the mutant STARD9-MD (R223A) could not bind to microtubules [Citation9].The results together suggested that STARD9-MD binds to interphase microtubules and indicated that STARD9 may be involved in regulating microtubule functions in the interphase cells.
Depletion of STARD9
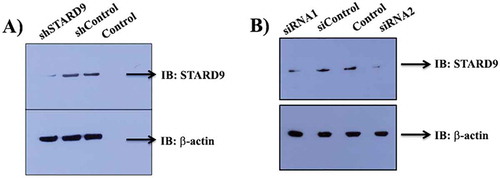
We have used two different strategies to knockdown STARD9 in HeLa cells. The target-specific shRNA and two different siRNAs were used to deplete STARD9 in cells (, Supplementary Table 1). shSTARD9, siRNA1 and siRNA2 treatment reduced the expression level of STARD9 by 95 ± 4, 70 and 72%, respectively. We used similar siRNA sequences to target STARD9 as reported earlier [Citation9]. The level of the depletion of STARD9 with siRNA was found to be similar to the previous study [Citation9]. A vector control for the shSTARD9 and a scrambled sequence for the siRNA did not reduce the expression level of STARD9 in HeLa cells. A partial rescue of the phenotype was observed upon the expression of STARD9-MD in HeLa cells indicating that the siRNA used was target specific [Citation9].
The depletion of STARD9 caused an extensive disassembly of the interphase microtubules
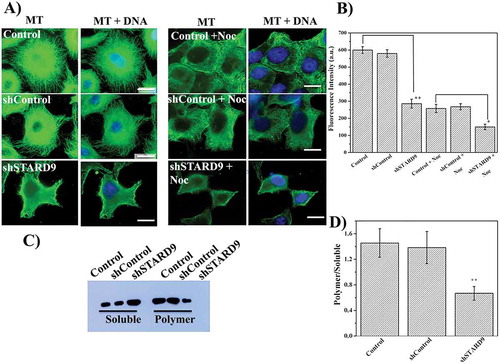
HeLa cells showed large numbers of distinct microtubules, well spread in the cytoplasm (, left panel). The shControl treated cells displayed intact microtubule filaments well spread in the cytoplasm similar to the control HeLa cells. However, microtubules were found to be visibly depolymerized in the STARD9-depleted cells as evident by the diffused staining of α-tubulin (, left panel and ). We also checked the effect of a low concentration (100 nM) of nocodazole on STARD9-depleted cells (), right panel, and ). The intensity of microtubules in the STARD9-depleted cells upon 100 nM nocodazole treatment was determined to be significantly lower than the control HeLa cells treated with nocodazole (p < 0.05) indicating that microtubules in the STARD9-depleted cells were more labile than microtubules in the control cells. Further, the polymeric and soluble fraction of microtubules from the control, shControl and shSTARD9 treated cells were quantified using western blots (). The ratio of polymeric to soluble tubulin decreased strongly in shSTARD9 depleted HeLa cells as compared to control and shControl treated HeLa cells. For example, the polymer/soluble tubulin ratio was determined to be 1.5 ± 0.4, 1.4 ± 0.5 and 0.7 ± 0.1 in control, shControl and shSTARD9 depleted HeLa cells, respectively suggesting that the depletion of STARD9 strongly depolymerized microtubules ().
Figure 2. STARD9 depletion with shRNA depolymerizes microtubules in HeLa cells. (a) HeLa cells were seeded on coverslips and transfection was performed without or with shControl or shSTARD9 with lipofectamine 3000 for 48 h. Further, immunostaining was performed with anti-tubulin IgG (left panel). In a separate experiment, the control and STARD9-depleted HeLa cells were incubated with 100 nM nocodazole for 2 h and immunostaining was done as mentioned above (right panel). Scale bar represents 10 μm. (b) The fluorescence intensity represents an average value obtained (n = 50 cells) from the region of interest (ROI). Error bar represents S.D. and p-value (** < 0.001; * < 0.05) was determined by student t-test. (c) HeLa cells were treated without or with shControl or shSTARD9 for 48 h and the cells were collected by centrifugation. The soluble fraction of tubulin was obtained by incubating the cell pellet with PEM buffer containing 0.5% triton X-100 and 25% glycerol for 2 min at 37°C. The polymeric tubulin was obtained by re-suspending the pellet in RIPA buffer for 1 h at 4°C. An equal amount of proteins was analyzed on SDS-PAGE and immuno-blotting was performed with anti-tubulin IgG. The experiment was performed 5 times. (d) The intensity of soluble and polymer tubulin was quantified with ImageJ and is plotted as an average of the ratio of polymer/soluble obtained from 5 independent set of experiments. Error bar represents S.D. and ** indicates p < 0.001.
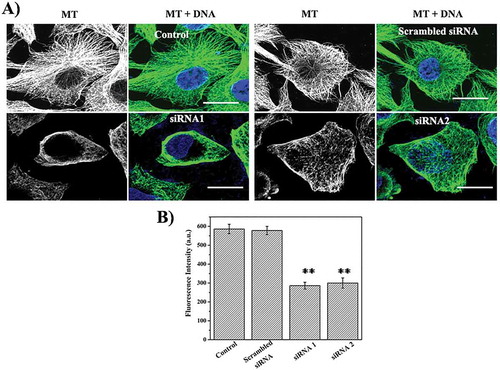
To examine the specificity of the effects of STARD9 on the interphase microtubules, STARD9 was also depleted using two different siRNAs. siSTARD9 depleted HeLa cells showed depolymerization of interphase microtubules (). However, scrambled siRNA treated HeLa cells displayed dense microtubule network similar to control HeLa cells ()). A significant reduction in the microtubule-network was found in both siRNA1 and siRNA2 treated HeLa cells as compared to the control and scrambled siRNA-treated HeLa cells ()). We also immunostained microtubules in control and STARD9-depleted HCT116 cells and observed a significant depolymerization of microtubules in HCT116 cells similar to the HeLa cells (Supplementary Figure3). The result together indicated that the depletion of STARD9 visibly depolymerizes microtubules and indicated that STARD9 may have a role in the stabilization of microtubules.
Figure 3. Depletion of STARD9 with siRNA depolymerizes microtubules in HeLa cells. (a) STARD9 depletion was performed with two different siRNAs (siRNA1 and siRNA2) and a scrambled siRNA with lipofectamine RNAiMAX for 48 h. Immunostaining was performed with anti-tubulin IgG. DNA staining was done with Hoechst 33258. Scale bar represents 10 µm. (b) The graph is plotted as an average microtubule intensity of 50 cells in case of control, siRNA1, siRNA2 or scrambled treated cells. The intensity of microtubule was determined using imageJ. ** indicates p < 0.001.
Over-expression of STARD9-MD in HeLa cells stabilized microtubules against nocodazole and cold treatments
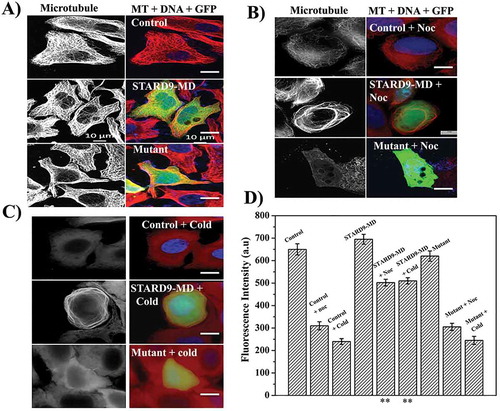
The cells showing green fluorescence signal were considered positive for STARD9-MD or mutant STARD9-MD (R223A) plasmid transfection. Non-transfected HeLa cells showed typical microtubules, well spread in the cytoplasm similar to that of the GFP-expressing HeLa cells (, Supplementary Figure4). However, microtubules intensity appeared to be increased in STARD9-MD transfected HeLa cells as compared to the GFP-STARD9-MD (R223A) and GFP transfected cells. The non-transfected and transfected (GFP-STARD9-MD, GFP-STARD9-MD (R223A) and GFP) HeLa cells were incubated with nocodazole (300 nM) for 1 h (, Supplementary Figure4). As expected, a significant depolymerization of microtubules was observed in control HeLa cells treated with 300 nM nocodazole. Although some microtubules were depolymerized in STARD9-MD transfected HeLa cells upon nocodazole treatment, microtubule bundles were also observed in the STARD9-MD transfected HeLa cells. Similar to the control HeLa cells, microtubules in GFP-transfected HeLa cells were strongly depolymerized upon nocodazole treatment (Supplementary Figure4). The mutant STARD9-MD transfected cells also showed microtubule depolymerization in the presence of nocodazole similar to the control and GFP transfected HeLa cells. We also treated the non-transfected and transfected HeLa cells with cold and observed microtubule bundles in STARD9-MD transfected cells ()). In contrast, microtubules were depolymerized in the case of control, GFP-STARD9-MD mutant (R223A) and GFP transfected HeLa cells (), Supplementary Figure4). The intensity of microtubules in STARD9-MD transfected HeLa cells were determined to be significantly higher than the control or STARD9-MD-(R223A) mutant cells when treated with either nocodazole or cold () (p < 0.001). The findings indicated that STARD9 may have a role in the stabilization of microtubules.
Figure 4. Expression of STARD9-MD stabilizes microtubules against nocodazole and cold. (a) HeLa cells were non-transfected or transfected with GFP-STARD9-MD or STARD9-MD R223A (mutant) for 24 h. The cells were fixed and immunostaining was performed with anti-α tubulin IgG. The control and transfected HeLa cells (indicated by arrows, GFP-STARD9-MD or STARD9-MD R223A) were incubated with (b) 300 nM nocodazole for 1 h or (c) cold (ice) for 30 min and processed for immunostaining with anti-tubulin IgG. DNA was stained with Hoechst 33258. Scale bar: 10 µm. (d) The average fluorescence intensity of microtubules from 50 cells was determined in case of control transfected HeLa cells with and without treatment with cold or nocodazole. Error bar indicates S.D. and p < 0.001(**).
The depletion of STARD9 suppressed microtubule dynamics in spindles
Both control and STARD9 depleted cells were synchronized with thymidine to increase the population of mitotic cells. Using fluorescent recovery after photo bleaching assay (FRAP) [Citation13,Citation14], we determined the effect of the depletion of STARD9 on spindle microtubule dynamics in live HeLa cells (, ). The recovery of the fluorescence intensity was monitored until the saturation value (). The control and shControl treated cells showed a recovery of fluorescence intensity up to 48 ± 5 and 47 ± 6%, respectively. In contrast, the shSTARD9-depleted cells showed only 10% recovery of the fluorescence intensity. As expected, no recovery of the GFP fluorescence occurred in the mitotic cells fixed with 4% formaldehyde. The fluorescence data were corrected for photobleaching rate and fitted into the first order exponential curve. The rate of recovery was determined to be 0.080 ± 0.02 and 0.079 ± 0.02 sec−1 in case of control and shControl treated HeLa cells, respectively (). In contrast, the rate constant for the STARD9-depleted HeLa cells was determined to be 0.034 ± 0.007 sec−1. The half recovery time (t1/2) was determined to be increased by 142% in the STARD9 depleted cells as compared to the control cells (). The results indicated that the depletion of STARD9 suppressed the dynamics of microtubules in the mitotic spindles.
Table 1. The rate constant and the half recovery time were determined from the analysis of fluorescent values fitted in the equation mentioned in method section using GraphPad Prism software. Shown are the average rate constant (k) and t1/2 values with ± as S.D. and n represents the number of cells scored in each case.
Figure 5. STARD9 depletion suppressed spindle microtubule dynamics in HeLa cells. (a) HeLa cells expressing GFP-α-tubulin were either treated without or with shControl or shSTARD9 for 36 h. Further, cells were blocked at interphase using 2 mM thymidine for 18 h. Five-hour post-release of thymidine fluorescent recovery after photobleaching (FRAP) was performed with the mitotic (bipolar) HeLa cells by bleaching a rectangular area with a high-intensity laser. Shown are the representative images obtained pre-bleach and post-bleach in case of control, shControl and shSTARD9 treated HeLa cells. Live cell imaging was done with confocal laser scanning microscope equipped with temperature (37°C) and CO2 (5%) control unit. Scale bar is shown in the figure. (b) The % fluorescence recovery of 15 cells in each experimental condition was plotted against time with ± as a standard deviation. (c) Control, shSTARD9, and shControl HeLa cells were fixed and immunostained with anti-γ-tubulin IgG and anti-α tubulin IgG. DNA was stained with Hoechst 33258. Scale bar shown in the figure. Shown is the maximum intensity projection of z-stacks in each case. (d) The graph is plotted as an average of the interpolar distance calculated from 30 cells in case of control, shControl and shSTARD9 depleted HeLa cells. Error bar indicates S.D. ** indicates p < 0.001.
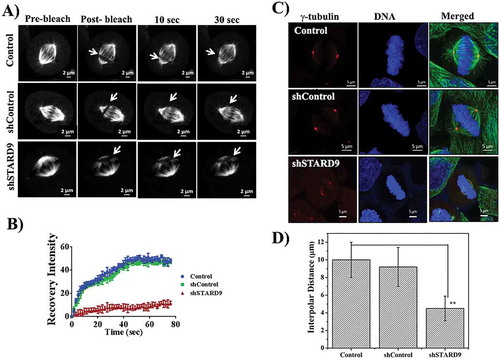
The distance between the two spindle poles in control, shControl or shSTARD9 treated HeLa cells was determined ()). The depletion of STARD9 in HeLa cells induced a mitotic block as reported earlier [Citation9]. We imaged metaphase cells (where the chromosomes were aligned at the equatorial plates with two gamma-tubulin foci and bipolar spindles) in control, shControl, and shSTARD9-depleted HeLa cells in the presence of MG132 (), Supplementary Figure 5). The interpolar distance was reduced by 55 ± 11% in the STARD9-depleted HeLa cells as compared to that of control and shControl treated cells indicating that STARD9 is involved in the formation of the bipolar mitotic spindle ()).
The depletion of STARD9 inhibited the reassembly of both interphase and spindle microtubules
Control and STARD9-depleted HeLa cells were incubated on ice for 30 min. The reassembly kinetics of microtubules was monitored by putting the cells in an incubator at 37°C. The cells were fixed at different time intervals and the growth of microtubules was estimated. Microtubules in both control and STARD9-depleted interphase cells were found to be strongly depolymerized upon cold treatment (Figure 6(a)). In control cells, the interphase microtubules were fully reassembled after 30 min while the interphase microtubules were not visibly formed in the STARD9-depleted HeLa cells even after 30 minutes of reassembly (). The fluorescence intensity of interphase microtubules in both STARD9-depleted and control HeLa cells was determined by monitoring 100 cells in each of the experimental conditions ()). The fluorescence intensity of microtubules was found to increase from 400 ± 10 to 910 ± 16 and 398 ± 8 to 460 ± 9 after 30 min of reassembly in control and STARD9-depleted HeLa cells, respectively (p < 0.001) suggesting that the depletion of STARD9 inhibited the reassembly of microtubules in HeLa cells ()). In a similar experiment, the depletion of STARD9 was found to strongly suppress the regrowth of spindle microtubules in HeLa cells (Figure 7). The fluorescence intensity of the mitotic spindles was found to increase from 378 ± 8 to 647 ± 30 and 352 ± 12 to 431 ± 11 after 30 min of spindle reassembly in control and STARD9-depleted HeLa cells, respectively (p < 0.01). The result also indicated that STARD9 is involved in the nucleation of mitotic microtubules ().
Figure 6. STARD9 depletion inhibited microtubule reassembly in interphase HeLa cells. (a) Microtubules were disassembled by incubating the cells on ice. The reassembly kinetics of the microtubules was monitored by incubating the cells for different (0, 5, 10, 15, 20, 25 and 30 min) durations at 37°C. The cells were fixed at the indicated time point and immunostaining was performed with anti-α tubulin IgG. Scale bar represents 10 µm. The experiment was performed twice. (b) The bar graph represents the fluorescence intensities of microtubules in control and STARD9-depleted HeLa cells fixed at different time points. In each time point, 50 cells were measured. The experiment was performed twice. ± indicates S.D., p < 0.001.
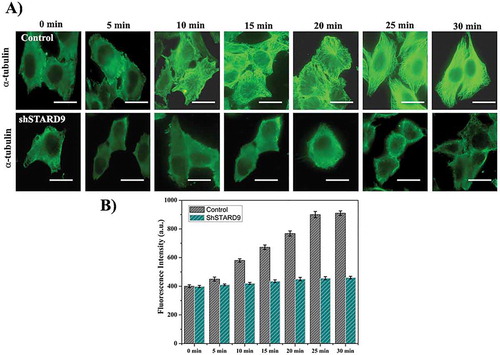
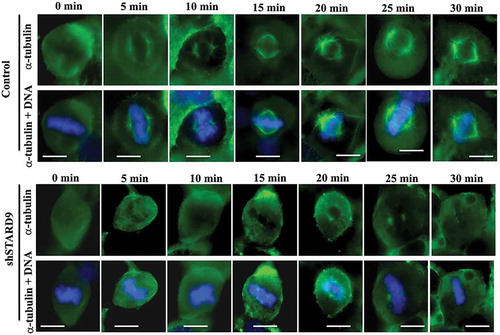
Figure 7. STARD9 depletion inhibited the reassembly of mitotic spindles. Shown are the representative mitotic cells in control and STARD9-depleted HeLa cells at different (0, 5, 10, 15, 20, 25 and 30 min) time intervals. Scale bar represents 10 µm. DNA was stained with hoechst 33258. The experiment was conducted two times.
STARD9 depletion inhibited chromosome separation
DNA and gamma-tubulin in STARD9-depleted HeLa cells were imaged using confocal microscopy (Supplementary Figure 6). The maximum intensity projection image was constructed from the z-stacks and the depletion of STARD9 was found to induce mitotic block with the cells at different stages of mitosis. A majority of the mitotic cells were found to be arrested at prometaphase stage with single, double or multiple gamma-tubulin foci. However, few of the mitotic cells were also found to have chromosomes aligned at the metaphase plate with two distinct gamma-tubulin foci and congression defects (Supplementary Figure6). Thus, the data suggested that the depletion of STARD9 significantly perturbed the organization of mitotic cells and induced defects in chromosome congression. We also analyzed the effect of the depletion of STARD9 on chromosome separation using mCherry-tagged H2B expressing HeLa cells (Supplementary Figure7A). The live cell imaging of control and shControl HeLa cells showed that the chromosomes were properly aligned at the equatorial plate and the chromosomes started separating towards the two poles showing anaphase (Supplementary Figure7A). The cells were successfully divided into two daughter cells (Supplementary Figure7A). The process of mitotic progression (mitosis to two daughter cells) was completed in ~ 60 min in both control and shControl-treated HeLa cells (Supplementary Figure7B). In contrast to the control cells, the separation of chromosomes was strongly inhibited in the STARD9-depleted prometaphase cells (Supplementary Figure7A and B).
STARD9 depletion synergized with vinblastine treatment
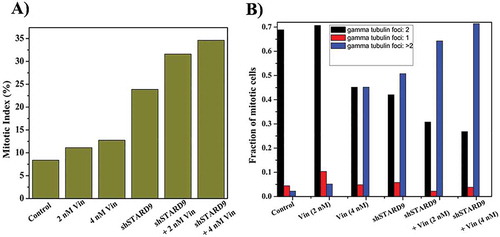
Since the depletion of STARD9 caused depolymerization of microtubules in HeLa cells, the effect of a well-known microtubule depolymerizing agent vinblastine was examined on STARD9-depleted cells. The half-maximal inhibitory concentration (IC50) for vinblastine in HeLa cells and STARD9-depleted HeLa cells was determined to be 4.1 ± 0.5 and 1.4 ± 0.2 nM, respectively (p < 0.05) ()). Further, we checked the effect of low concentrations (2 and 4 nM) of vinblastine on the microtubules in control and STARD9-depleted HeLa cells ()). Low concentrations of vinblastine produced much stronger depolymerization of microtubules in the STARD9-depleted HeLa cells compared to the control cells ()). The percentage mitotic index in HeLa cells was determined to be 8, 11, and 13 in the absence and presence of 2 and 4 nM vinblastine, respectively while it was found to be 24% in the STARD9-depleted HeLa cells (, Supplementary Figure 8). The mitotic index was determined to be 32 and 34% in the STARD9-depleted HeLa cells in the presence of 2 and 4 nM vinblastine, respectively (). Further, we quantified the number of gamma-tubulin foci in case of control and STARD9 depleted HeLa cells in the presence of vinblastine (, Supplementary Figure8). In control cells, 76% of the total mitotic cells had two gamma-tubulin foci. In 2 and 4 nM vinblastine-treated HeLa cells, 70 and 45% of the mitotic cells were found to have two gamma-tubulin foci respectively. In contrast, 42, 30 and 27% of the cells displayed double gamma-tubulin foci in STARD9-depleted HeLa cells in the absence and presence of 2 and 4 nM vinblastine, respectively. STARD9 depleted HeLa cells showed 50% of the cells with multipolar foci. A significant increase in the multipolar cells was found when STARD9-depleted cells were treated with vinblastine. For example, 64 and 71% of the mitotic cells were found to be multipolar in STARD9-depleted HeLa cells treated with 2 and 4 nM vinblastine, respectively (), Supplementary Figure8).The finding suggested that the depletion of STARD9 synergizes with vinblastine treatment and increases cell death by forming multipolar cells.
Figure 8. Vinblastine treatment produced a strong anti-proliferative effect in STARD9-depleted HeLa cells. (a) STARD9 depletion was done in HeLa cells for 36 h with shSTARD9. The control (●) and shSTARD9 depleted (■) HeLa cells were incubated with different concentrations (78, 156, 310, 625, 1250, 2500, 5000, 10000, 20000, 40000 pM) of vinblastine for 24 h. The inhibition of cell proliferation was determined by sulforhodamine B assay. The average value determined from three independent sets of experiments is provided. Error bars indicate S.D., p < 0.05. (b) Control and STARD9-depleted HeLa cells were incubated without and with vinblastine (2 and 4 nM) for 10 h and processed for immunostaining with anti-α-tubulin IgG. DNA was stained with Hoechst 33258 and scale bar represents 10 μm.
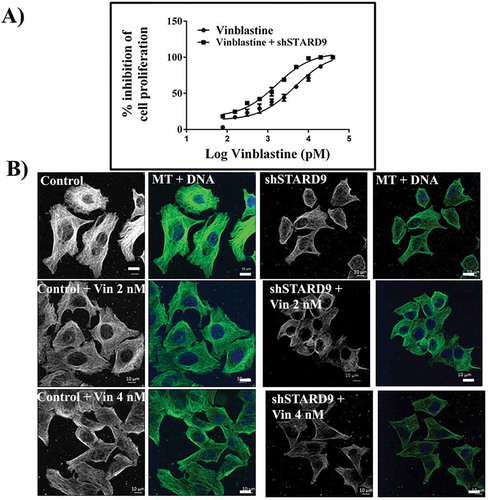
Figure 9. Vinblastine treatment increased mitotic index and multipolar cells in STARD9-depleted HeLa cells. (a) HeLa cells were treated without or with shControl or shSTARD9 for 36 h. Subsequently, the cells were incubated without or with vinblastine (2 and 4 nM) for 10 h. Immunostaining was performed with anti-γ-tubulin IgG. The chromosomes were stained with Hoechst 33258. Further, the mitotic index (ratio of the number of cells at mitosis/total number of cells × 100) was determined in each case. The number of cells scored was 500 for each of the experimental conditions. (b) Number of cells with single, double or multiple gamma-tubulin foci were scored and plotted as a fraction of total mitotic cells. A total number of mitotic cells taken for calculation were 250 in each case. The experiment was performed twice.
Discussion
In the present study, we have elucidated the role of a centrosomal protein, STARD9, on the assembly and stability of microtubules. The depletion of STARD9 strongly suppressed the reassembly of spindle and interphase microtubules indicating a role of STARD9 in the nucleation of microtubules. The depletion of STARD9 induced microtubule depolymerization in HeLa cells and the over-expression of the motor domain of STARD9 (STARD9-MD) stabilized microtubules in response to cold and nocodazole treatment. In addition, STARD9 depletion dampened the dynamics of microtubules in the mitotic spindles suggesting that STARD9 regulates tubulin exchange in the mitotic spindles. Further, STARD9-depleted cells showed increased sensitivity towards vinblastine suggesting that the combined effect of STARD9 depletion and anti-tubulin drug could be used as a strategy for cancer treatment.
STARD9 regulates microtubule assembly and spindle dynamics
STARD9-MD co-localized along the length of microtubules in HeLa cells and co-immunoprecipitated with tubulin. STARD9 was also precipitated from the polymeric fraction of tubulin with anti-tubulin IgG, suggesting that STARD9 binds to the microtubules in HeLa cells. In an earlier report [Citation9], the effect of siRNA-mediated depletion of STARD9 was partially rescued by expressing STARD9-MD in HeLa cells. In this study, we found that the depletion of STARD9 with shRNA or siRNA significantly reduced the level of microtubules in HeLa cells implying the role of STARD9 in promoting microtubule assembly or stability. The transfected STARD9-MD HeLa cells were shown to form microtubule bundles indicating that STARD9 could either induce microtubule polymerization or enhance the stability of the existing microtubules or both. Further, microtubules in STARD9-MD transfected cells showed resistance to nocodazole and cold treatment than the control cells indicating that STARD9 can stabilize microtubules. STARD9 also appears to regulate the nucleation of microtubules as the depletion of STARD9 strongly inhibited the re-growth of both spindle and interphase microtubules in HeLa cells. In this work, STARD9 was co-precipitated with tubulin from the polymerized fraction of tubulin and STARD9-MD was found to decorate microtubules in HeLa cells. In addition, purified STARD9-MD was found to bind microtubules [Citation9]. The results together suggested that STARD9 binds to microtubules and the binding of STARD9 to microtubules increases the assembly and stability of microtubules.
The FRAP experiment showed that the depletion of STARD9 decreased tubulin turnover suggesting that STARD9 may be involved in tubulin addition to the microtubule ends causing increased growth rate. Recently, kinesin-5 was reported to increase the incorporation of tubulin dimers at the plus end of the microtubules and thereby, to promote microtubule polymerization [Citation15]. A centromere protein CENP-E was reported to induce microtubule elongation by the addition of tubulin dimers to the microtubule ends in an ATP-dependent manner [Citation16]. We found that the ectopic expression of STARD9-MD induced the formation of fibrous microtubule bundles near the nucleus similar to the expression of the C-terminal domain of Cep57 [Citation17]. Further, the expression of STARD9-MD was found to induce nocodazole resistant and cold resistant microtubule bundles in HeLa cells similar to the resistant microtubule bundles formed in Cos-7 cells upon expression of Myc-CLAMP [Citation18]. These observations together suggested that ectopic expression of STARD9-MD can induce the bundling of microtubules in HeLa cells. The formation of bundles in the STARD9-MD transfected cells suggested that STARD9 can increase the lateral interactions between the microtubule protofilaments. STARD9, being a large size protein could bind to two different protofilaments and enhance the lateral interactions between them. Similar to kinesin-12 [Citation19], single or multiple STARD9 could crosslink the microtubule plus end leading to the formation of parallel bundles or like kinesin-5 motor [Citation20], STARD9 could crosslink anti-parallel microtubules.
A perturbation in the spindle dynamics is reported to induce the formation of disoriented mitotic spindles [Citation21]. Similar to the knockdown of other microtubule stabilizing plus end binding proteins like EB1 [Citation22], Msps [Citation23] or Mast [Citation24], STARD9 depletion also induced the formation of spindles with reduced interpolar distance [Citation21,Citation25] indicating that STARD9 could also regulate spindle dynamics. The FRAP analysis suggested that the depletion of STARD9 increased the value of t1/2 by 142%. A similar increase in t1/2 (approximately 1.3 times of control) was observed in case of knockdown of Mast protein in S2 cells, implying a decrease in tubulin turnover [Citation13]. In PtK-T-cells, a decrease of 75% in the t1/2 (from 7 ± 3 to 4 ± 2 sec) value was observed in the presence of a microtubule-stabilizing agent, taxol (40 nM), suggesting an increased assembly rate [Citation14]. Thus STARD9 could increase tubulin turnover in the mitotic spindle leading to effective cell division. The suppression of spindle microtubule dynamics in STARD9 depleted cells could impair the force generation at the spindle pole and thereby, inhibited the mitotic progression. STARD9-depleted HeLa cells which were arrested at different phases of mitosis failed to separate the chromosomes suggesting that STARD9 also has a role in the separation of chromosomes.
STARD9 as an anti-cancer target
Though microtubule-targeting drugs are widely used in cancer therapy, several drawbacks including neurological toxicity, development of resistance or neutropenia limit their utility [Citation26]. This led to the identification of a new class of agents, which target proteins such as the kinesins and kinases that are involved in mitosis. For example, inhibitors of kinesin spindle protein Eg5 (such as SB-715992,MK-0731) [Citation27,Citation28], Plk1 (Polo-like kinase 1) (BI 2536, ON01910) [Citation29,Citation30] and aurora kinase (MK-0457/VX-680) [Citation31] are in different stages of clinical trials. STARD9 could also emerge as a potent target for cancer therapy as the inhibition of STARD9 functions leads to an increased mitotic index with multipolar spindles in cancer cells. Intriguingly, the depletion of STARD9 was found to produce more pronounced effects in cancer cells (for example HeLa, HCT116, 460 M395, U2OS) than in the non-cancerous cells such as MCF10A, RPE or Lymphoblast cells [Citation9]. The STARD9 depleted cancer cells further undergo apoptosis-mediated cell death. Thus, the identification of compounds targeting STARD9 opens a new avenue for anti-mitotic mediated cancer therapeutics. The use of a drug in combination with the depletion of STARD9 can be an alternative therapeutic approach. For example, taxanes are combined with anthracycline or platinum analogs for the treatment of breast cancer and non-small cell lung cancer respectively [Citation32,Citation33]. STARD9 depleted HeLa cells were more sensitive to the vinblastine treatment as compared to normal HeLa cells. Vinblastine treatment increased the cytotoxic effect in the STARD9-depleted HeLa cells by increasing the number of cells with multipolar spindles. The findings suggest that the combination of STARD9 inhibitors (small molecules or RNAi) and a microtubule-depolymerizing agent (such as vinblastine) could be a future mode of cancer therapy.
Materials
Mouse monoclonal anti-tubulin IgG (T6074, Sigma), mouse monoclonal anti-γ-tubulin IgG (T5326, Sigma), FITC conjugated anti-rabbit IgG (F0382, Sigma), FITC conjugated anti-mouse IgG (F9137, Sigma). Vinblastine, nocodazole and thymidine were also procured from Sigma Aldrich (St Louis, MO, USA). Lipofectamine RNAimax, Lipofectamine 3000, fetal bovine serum (FBS), Opti-MEM reduced serum-free medium and anti-mouse Alexa 568 (A11005) were purchased from Invitrogen (Eugene, USA). The Super Signal West Pico Chemiluminescent Substrate was obtained from Thermo Scientific (Rockford, U.S.A.). STARD9 shRNA (catalog no.sc-63083-SH), Control shRNA plasmid A (catalog number: sc 108060), Protein A agarose beads STARD9 IgG, cyclin B1 and GAPDH (IgG) were purchased from Santa Cruz (California). The siRNAs were synthesized by Eurogentec (North America).
Cell culture
HeLa cells (cervical carcinoma) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, HiMedia) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% (v/v) antibiotic-antimycotic solution. HCT116 (colorectal carcinoma) cells were maintained in RPMI 1640 media with 10% FBS and 1% antibiotic-antimycotic solution. The cells were purchased from National Centre for Cell Science, Pune, India. HeLa cells expressing histone 2B-mCherry was a kind gift from Tapas Manna, IISER Thiruvananthapuram, India. The cell lines were authenticated using STR analysis and were found to be free from mycoplasma. The H2B cells were maintained in DMEM with 10% FBS and 1% antibiotic-antimycotic solution. The cells were maintained at 37°C in a humidified CO2 (5%) chamber.
Immunoprecipitation assay
We separated the polymeric and soluble fraction of tubulin [Citation34] and quantified the amount of STARD9 associated with the polymeric and soluble fraction of tubulin through western blot. For immunoprecipitation assay [Citation35], tubulin IgG and GAPDH IgG was incubated with the polymeric fraction of tubulin (input) followed by incubation with Protein A agarose beads for 3 h beads were then washed 3 times with PBS and proteins were eluted by boiling the beads with SDS loading buffer. The proteins were separated by SDS-PAGE and immuno-blotting was performed with anti-STARD9 IgG and anti-GAPDH IgG antibody. The experiment was performed thrice in each case.
STARD9 protein level in interphase and mitotic cells
We checked the presence of STARD9 in interphase and mitotic cells. The cells were synchronized in interphase and mitotic phase using thymidine and nocodazole, respectively [Citation36]. Briefly, HeLa cells (106cells/mL) were seeded in a flask and incubated with 2 mM thymidine for 18 h. For mitotic cells, HeLa cells were treated with 300 nM nocodazole for 14 h. The cells were collected through mitotic shake off [Citation36]. The interphase and mitotic cells were lysed and immuno-blotting was performed with anti-STARD9 IgG, anti-cyclin B1 IgG and anti-GAPDH IgG. The experiment was repeated 3 times.
STARD9-MD co-localizes with microtubules
The GFP-STARD9-MD (motor domain of STARD9) and GFP-STARD9-MD (R223A) mutant plasmids were a kind gift from Jorge Z. Torres (University of California, USA). HeLa cells seeded on polylysine-coated coverslips were transfected with GFP, GFP-STARD9-MD and GFP-STARD9-MD (R223A) mutant plasmids. The cells were then fixed with 4% formaldehyde and permeablized using 0.25% Triton X-100. Immunostaining was performed with anti-mouse GFP IgG and anti-rabbit tubulin IgG. Fluorescent tagging was done with anti-mouse FITC IgG and anti-rabbit Alexa 594. For immunoprecipitation [Citation35], the cell extract prepared from GFP-STARD9-MD transfected HeLa cells were incubated with anti-tubulin IgG and processed for immuno-blotting with anti-GFP IgG and anti-GAPDH IgG as described earlier. The experiment was done twice.
Depletion of STARD9
STARD9 was depleted in HeLa and HCT116 cells using shRNA (now onwards will be referred as shSTARD9) and control shRNA plasmid A (shControl). HeLa cells were seeded to 70% confluency and transfected with either shSTARD9 or shControl using Lipofectamine 3000. After the transfection, cells were incubated for 48 h. The cells were collected by centrifugation and lysed to collect the cell extract. The cell lysate was separated on 6% SDS-PAGE and immuno-blotted with anti- STARD9 and anti-actin IgG. The experiment was performed thrice. Similarly, the depletion of STARD9 in HeLa cells was also done with two different siRNAs (sequences similar to Torres, et. al [Citation9]) and a scrambled sequence using Lipofectamine RNAiMAX for 48 h. The experiment was performed twice. The target sequences for the siRNA and shRNA are shown in Supplementary Table 1. siRNA 1 and siRNA 2 were designed to target two different regions of STARD9 mRNA sequence. The scrambled sequence is a random non-specific sequence (Supplementary Table 1).
Effects of STARD9 depletion on interphase and mitotic HeLa cells
HeLa cells (80,000 cells/mL) were seeded on polylysine-coated coverslips and STARD9 depletion was performed using shRNA, siRNAs or scrambled siRNA as described earlier. Immunostaining was performed with anti-α tubulin IgG and the DNA was stained with Hoechst 33258 (10 µg/mL). In a separate experiment, 100 nM nocodazole was added to control; shControl or shSTARD9 treated cells for 2 h and further processed for immunostaining. The fluorescence intensity of 50 cells in experimental condition was quantified using ImageJ. For the quantification of fluorescence intensity, a region of interest was drawn outside the whole cell. STARD9 was also depleted in HCT116 cells using shSTARD9.
Quantification of soluble and polymer mass in STARD9 depleted cells
The extent of soluble and polymeric tubulin was quantified by separating the soluble and insoluble tubulin fraction [Citation37]. Briefly, control, shControl and shSTARD9 treated cells were collected by centrifugation and washed with PBS. Further, PEM buffer (50 mM PIPES, 3 mM MgCl2 and 1 mM EGTA) containing 0.5% Triton-X-100 and 25% glycerol was added to the cell pellet for 2 min to collect soluble tubulin at 37°C. Insoluble tubulin (polymerized MT) was collected by treating the pellet with RIPA buffer containing protease inhibitors for 1 h at 4°C. An equal amount of protein was then analyzed by western blotting with anti α-tubulin IgG. The intensity of tubulin was quantified with ImageJ and the ratio of polymer/soluble was determined. The experiment was done 5 times. In a separate experiment, both polymeric and soluble fraction of tubulin was separated on SDS-PAGE and western blotting was performed with anti-STARD9 IgG. The experiment was done thrice.
Effects of overexpression of STARD9-MD and its mutant on HeLa cells
HeLa cells were transfected without and with GFP-STARD9-MD, GFP-STARD9-MD (R223A) (mutant) and GFP plasmid using Lipofectamine 3000 for 24 h and immunostained with anti α-tubulin IgG as described above. In a separate experiment, HeLa cells were transfected with GFP-STARD9-MD, GFP-STARD9-MD (R223A) (mutant) and GFP and then, either incubated with nocodazole 300 nM (1 h) or provided cold treatment for 30 min. Microtubules were immunostained with anti-α-tubulin IgG and DNA was stained with Hoechst 33258. Microtubule intensity of 50 cells in each case was determined using ImageJ as mentioned before.
Measurement of spindle microtubule dynamics by fluorescence recovery after photobleaching in live HeLa cells
HeLa cells were transiently transfected with GFP-tagged α-tubulin plasmid using Lipofectamine 3000 for 24 h. Further, cells were synchronized in interphase using 2 mM thymidine treatment for 18 h [Citation36]. The cells were thoroughly washed with DPBS for thymidine release. Post 5 h of thymidine release, fluorescence recovery after photobleaching (FRAP) assay was performed. For shSTARD9 depleted HeLa cells, the depletion was performed using shRNA or shControl for 36 h. Subsequently, the cells were incubated with 2 mM thymidine for 18 h, carefully washed with DPBS, and the FRAP analysis was performed 5 h after releasing thymidine. STARD9 depletion induced abnormalities in spindle formation as most of the cells were multipolar. HeLa cells oriented in bipolar spindles were used for the FRAP analysis. A rectangular region of approximate area 3 × 5 µm2 between the poles and equatorial region were bleached using high power laser. The bleaching was done until the fluorescence intensity was decreased to ~ 70% [Citation13,Citation14]. Post-bleaching, the images were captured at an interval of 1.6 s till the fluorescence recovery value reached a plateau using a confocal laser scanning microscope with 63x oil immersion objective equipped with temperature and CO2 controller unit. Further, the fluorescence value was exported to Microsoft Excel and normalized fluorescence intensity was calculated using the following formula,
; where, Fs is the fluorescence intensity of the region of interest, Fb is the fluorescence intensity of background and r is the rate of photobleaching
. Fco is the fluorescence intensity at time zero and Fc is the fluorescence intensity at time t. The normalized fluorescence intensity values were fitted to one phase exponential decay equation using GraphPad Prism Software,
; where F0 is the F-value at time zero, plateau is the F value at infinite time and K is the rate constant [Citation38]. The percentage of fluorescence recovery was calculated by using the formula,
, where Fr is the fluorescence intensity at time t, Fb is the fluorescence intensity before photobleaching and Fa is the fluorescence intensity after photobleaching [Citation39].
Interpolar distance measurement
To determine the interpolar distance, the control, shControl, and shSTARD9 depleted HeLa cells were incubated with MG132 for 1h. Further, the cells were fixed and the immunostaining was performed with gamma-tubulin IgG and tubulin IgG. DNA was stained with Hoechst 33258. The images were captured in a confocal microscope with z-stacking and the maximum intensity projection of z-stacks was constructed. The interpolar distance of 30 cells in case of control, shControl, and shSTARD9 cells was determined using the Zen software.
Microtubule reassembly in STARD9 depleted HeLa cells
STARD9-depleted and control HeLa cells were incubated on ice for 30 min. Further, both control and STARD9-depleted HeLa cells were incubated with warm media and shifted to a carbon dioxide incubator at 37°C. The cells were fixed at 0, 5, 10, 15, 20, 25 and 30 min after incubation with warm media. Immunostaining was performed with anti-α tubulin IgG and DNA was stained with Hoechst 33258. The microtubule fluorescence intensity of 50 cells was determined in each case from the whole cell using ImageJ. The experiment was performed twice.
Chromosome separation in H2B-mcherry expressing HeLa cells
The stable H2B-mCherry expressing HeLa cells were used to study the chromosome separation in control and STARD9-depleted HeLa cells. STARD9 depletion was accomplished using shSTARD9 or shControl for 48 h. Further, control, shSTARD9 or shControl H2B cells were imaged using a spinning disk confocal microscope (Yokogawa CSU-X1) equipped with a temperature controller (37°C) and CO2 (5%) control unit with 40x oil immersion objectives. The images of the cells at the beginning of mitosis were captured for 3 h at an interval of 5 min. The images were processed and the distance between the sister’s chromosomes was calculated using the Zen software. The chromosome separation of 10 cells in each case was monitored.
Effects of STARD9 depletion in combination with vinblastine on HeLa cells
HeLa cells (50,000 cells/mL) were seeded in 96 well plates. STARD9 depletion was performed in HeLa cells using shSTARD9 for 36 h. HeLa cells and STARD9 depleted HeLa cells were grown without or with different concentrations (78, 156, 310, 625, 1250, 2500, 5000, 10000, 20000, 40000 pM) of vinblastine for 24 h. The half-maximal inhibitory concentration (IC50) of vinblastine in HeLa cells and STARD9-depleted HeLa cells was determined using the sulforhodamine B assay [Citation40]. The experiment was performed thrice. For immunostaining, the control and STARD9-depleted HeLa cells were incubated without and with vinblastine (2 and 4 nM) for 10 h. The cells were fixed and processed for immunostaining with anti-α tubulin IgG. DNA was stained with Hoechst 33258.
Mitotic index calculation
The cells were incubated without and with vinblastine (2 and 4 nM) for 10 h and immunostained with anti-γ-tubulin IgG. DNA was stained with Hoechst 33258. The fraction of cells with single, double and multiple gamma-tubulin foci were scored by counting 500 cells. Mitotic index was calculated as the percentage of mitotic cells to the total number of cells. The experiment was done twice and in each case, 250 cells were scored.
Microscopy data
All the images were captured in a laser scanning confocal microscope (Zeiss Axio-Observer Z1) with Plan-Apochromat 63x/1.4 NA (oil) objective lens unless mentioned.
Statistical analysis
The statistical analysis was performed, where ± indicates the standard deviation (S.D.). The significance value was determined using Student’s t-test, p-value <0.05 was considered to be significant.
Author contributions
SS and DP designed experiments and analyzed data. SS performed the experiments. SS and DP contributed to the manuscript preparation.
Supplemental Material
Download MS Word (17.8 MB)Acknowledgments
We thank Prof. Jorge Z. Torres (University of California, USA) for providing us GFP-STARD9-MD and GFP-STARD9-MD (R223A) mutant plasmids. We also thank IIT Bombay for providing central facilities like LSM confocal microscope, spinning disk confocal microscope, cell culture. SS thanks University grant commission, Government of India for a doctoral fellowship. We thank Anuradha Kumari for her support and critical suggestions. This work was supported by a grant [no. BT/PR14618/BRB/10/1418/2015] from the Department of Biotechnology, Government of India. DP is supported by TATA Innovation Fellowship, Department of Biotechnology, Government of India.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data can be accessed here.
Additional information
Funding
References
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002 Feb;14(1):25–34. PubMed PMID: 11792541.
- Cassimeris L, Morabito J. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol Biol Cell. 2004 Apr;15(4):1580–1590. PubMed PMID: 14718566; PubMed Central PMCID: PMC379257.
- Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002 Oct 28;159(2):255–266. PubMed PMID: 12403812; PubMed Central PMCID: PMC2173044.
- Kumari A, Panda D. Regulation of microtubule stability by centrosomal proteins. IUBMB Life. 2018 Jul;70(7):602–611. PubMed PMID: 29734495.
- Mahen R, Venkitaraman AR. Pattern formation in centrosome assembly. Curr Opin Cell Biol. 2012 Feb;24(1):14–23. PubMed PMID: 22245706.
- Oshimori N, Ohsugi M, Yamamoto T. The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat Cell Biol. 2006 Oct;8(10):1095–1101. PubMed PMID: 16980960.
- Venghateri JB, Jindal B, Panda D. The centrosome: a prospective entrant in cancer therapy. Expert Opin Ther Targets. 2015 Jul;19(7):957–972. PubMed PMID: 25787715.
- Halama N, Grauling-Halama SA, Jager D. Identification and characterization of the human StARD9 gene in the LGMD2A-region on chromosome 15q15 by in silico methods. Int J Mol Med. 2006 Oct;18(4):653–656. PubMed PMID: 16964419.
- Torres JZ, Summers MK, Peterson D, et al. The STARD9/Kif16a kinesin associates with mitotic microtubules and regulates spindle pole assembly. Cell. 2011 Dec 09;147(6):1309–1323. PubMed PMID: 22153075; PubMed Central PMCID: PMC4180425.
- Senese S, Cheung K, Lo YC, et al. A unique insertion in STARD9’s motor domain regulates its stability. Mol Biol Cell. 2015 Feb 01;26(3):440–452. PubMed PMID: 25501367; PubMed Central PMCID: PMC4310736.
- De Luca M, Brunetto L, Asteriti IA, et al. Aurora-A and ch-TOG act in a common pathway in control of spindle pole integrity. Oncogene. 2008 Nov 20;27(51):6539–6549. PubMed PMID: 18663358.
- Lawo S, Bashkurov M, Mullin M, et al. HAUS, the 8-subunit human augmin complex, regulates centrosome and spindle integrity. Current Biology: CB. 2009 May 26;19(10):816–826. PubMed PMID: 19427217.
- Buster DW, Zhang D, Sharp DJ. Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol Biol Cell. 2007 Aug;18(8):3094–3104. PubMed PMID: 17553931; PubMed Central PMCID: PMC1949370.
- Rizk RS, Bohannon KP, Wetzel LA, et al. MCAK and paclitaxel have differential effects on spindle microtubule organization and dynamics. Mol Biol Cell. 2009 Mar;20(6):1639–1651. PubMed PMID: 19158381; PubMed Central PMCID: PMC2655246.
- Chen Y, Hancock WO. Kinesin-5 is a microtubule polymerase. Nat Commun. 2015 Oct 06;6:8160.
- Sardar HS, Luczak VG, Lopez MM, et al. Mitotic kinesin CENP-E promotes microtubule plus-end elongation. Current Biology: CB. 2010 Sep 28;20(18):1648–1653. PubMed PMID: 20797864; PubMed Central PMCID: PMC2946434.
- Momotani K, As K, Miyake T, et al. Cep57, a multidomain protein with unique microtubule and centrosomal localization domains. Biochem J. 2008 Jun 1;412(2):265–273. PubMed PMID: 18294141; PubMed Central PMCID: PMC4351815.
- Dougherty GW, Adler HJ, Rzadzinska A, et al. CLAMP, a novel microtubule-associated protein with EB-type calponin homology. Cell Motil Cytoskeleton. 2005 Nov;62(3):141–156. PubMed PMID: 16206169.
- Drechsler H, McAinsh AD. Kinesin-12 motors cooperate to suppress microtubule catastrophes and drive the formation of parallel microtubule bundles. Proc Natl Acad Sci U S A. 2016 Mar 22;113(12):E1635–44. PubMed PMID: 26969727; PubMed Central PMCID: PMC4812750.
- Sharp DJ, McDonald KL, Brown HM, et al. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of drosophila embryonic mitotic spindles. J Cell Biol. 1999 Jan 11;144(1):125–138. PubMed PMID: 9885249; PubMed Central PMCID: PMC2148119.
- Goshima G, Wollman R, Stuurman N, et al. Length control of the metaphase spindle. Current Biology: CB. 2005 Nov 22;15(22):1979–1988. PubMed PMID: 16303556.
- Rogers SL, Rogers GC, Sharp DJ, et al. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002 Sep 02;158(5):873–884. PubMed PMID: 12213835; PubMed Central PMCID: PMC2173155.
- Brittle AL, Ohkura H. Mini spindles, the XMAP215 homologue, suppresses pausing of interphase microtubules in Drosophila. EMBO J. 2005 Apr 06;24(7):1387–1396. PubMed PMID: 15775959; PubMed Central PMCID: PMC1142550.
- Maiato H, Sampaio P, Lemos CL, et al. MAST/Orbit has a role in microtubule-kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J Cell Biol. 2002 May 27;157(5):749–760. PubMed PMID: 12034769; PubMed Central PMCID: PMC2173411.
- Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Current Biology: CB. 2009 Sep 15;19(17):R749–61. PubMed PMID: 19906577; PubMed Central PMCID: PMC2791830.
- Jackson JR, Patrick DR, Dar MM, et al. Targeted anti-mitotic therapies: can we improve on tubulin agents? Nat Rev Cancer. 2007 Feb;7(2):107–117. PubMed PMID: 17251917.
- Blagden SP, Molife LR, Seebaran A, et al. A phase I trial of ispinesib, a kinesin spindle protein inhibitor, with docetaxel in patients with advanced solid tumours. Br J Cancer. 2008 Mar 11;98(5):894–899. PubMed PMID: 18319713; PubMed Central PMCID: PMC2266864.
- Holen K, DiPaola R, Liu G, et al. A phase I trial of MK-0731, a kinesin spindle protein (KSP) inhibitor, in patients with solid tumors. Invest New Drugs. 2012 Jun;30(3):1088–1095. PubMed PMID: 21424701; PubMed Central PMCID: PMC3394096.
- Hofheinz RD, Al-Batran SE, Hochhaus A, et al. An open-label, phase I study of the polo-like kinase-1 inhibitor, BI 2536, in patients with advanced solid tumors. Clin Cancer Res. 2010 Sep 15;16(18):4666–4674. PubMed PMID: 20682708.
- Jimeno A, Li J, Messersmith WA, et al. Phase I study of ON 01910.Na, a novel modulator of the polo-like kinase 1 pathway, in adult patients with solid tumors. J Clinical Oncology: Official Journal Am Soc Clin Oncol. 2008 Dec 01;26(34):5504–5510. PubMed PMID: 18955447; PubMed Central PMCID: PMC4824307.
- Traynor AM, Hewitt M, Liu G, et al. Phase I dose escalation study of MK-0457, a novel aurora kinase inhibitor, in adult patients with advanced solid tumors. Cancer Chemother Pharmacol. 2011 Feb;67(2):305–314. PubMed PMID: 20386909; PubMed Central PMCID: PMC3050703.
- Jones S, Hainsworth J, Burris HA 3rd, et al. Phase I study of JM-216 (an oral platinum analogue) in combination with paclitaxel in patients with advanced malignancies. Invest New Drugs. 2002 Feb;20(1):55–61. PubMed PMID: 12008664.
- Nabholtz JM, Riva A. Taxane/anthracycline combinations: setting a new standard in breast cancer? Oncologist. 2001;6(Suppl 3): 5–12. PubMed PMID: 11346678.
- Ochoa CD, Stevens T, Balczon R. Cold exposure reveals two populations of microtubules in pulmonary endothelia. Am J Physiol Lung Cell Mol Physiol. 2011 Jan;300(1):L132–8. PubMed PMID: 20971804; PubMed Central PMCID: PMC3023290.
- Free RB, Hazelwood LA, Sibley DR. Identifying novel protein-protein interactions using co-immunoprecipitation and mass spectroscopy. Curr Protoc Neurosci. 2009 Jan; Chapter 5: Unit5 28. PubMed PMID: 19170023; PubMed Central PMCID: PMC4752115. DOI:10.1002/0471142301.ns0528s46.
- Ht M, Ry P. Synchronization of HeLa cells. Methods Mol Biol. 2011;761:151–161. PubMed PMID: 21755447.
- Sidhaye VK, Chau E, Srivastava V, et al. A novel role for aquaporin-5 in enhancing microtubule organization and stability. PloS One. 2012;7(6):e38717. PubMed PMID: 22715407; PubMed Central PMCID: PMC3371026.
- Zheng CY, Petralia RS, Wang YX, et al. Fluorescence recovery after photobleaching (FRAP) of fluorescence tagged proteins in dendritic spines of cultured hippocampal neurons. Journal Vis Exp: JoVE. 2011 Apr 16; (50): PubMed PMID: 21525845; PubMed Central PMCID: PMC3339873. doi: 10.3791/2568.
- Giese B, Au-Yeung CK, Herrmann A, et al. Long term association of the cytokine receptor gp130 and the janus kinase Jak1 revealed by FRAP analysis. J Biol Chem. 2003 Oct 3;278(40):39205–39213. PubMed PMID: 12878601.
- Skehan P, Storeng R, Scudiero D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990 Jul 04;82(13):1107–1112. PubMed PMID: 2359136.