Abstract
Connexin oligomerization and trafficking are regulated processes. To identify proteins that control connexin43 (Cx43), a screen was designed using HeLa cells expressing a Cx43 construct with di-lysine endoplasmic reticulum (ER)-retention/retrieval motif, Cx43-HKKSL. At moderate levels of expression, Cx43-HKKSL is retained in the ER as monomers; however, Cx43-HKKSL stably overexpressed by HeLa cells localizes to the perinuclear region and oligomerizes. HeLa/Cx43-HKKSL overexpressors were transiently transfected with pooled clones from a human kidney cDNA library and used immunofluorescence microscopy to identify cDNAs that enabled overexpressed Cx43-HKKSL to convert from a perinuclear to ER localization pattern. Using this approach, a small molecular weight GTPase, rab20, was identified as a candidate protein with the ability to regulate Cx43 trafficking. Enhanced green florescent protein (EGFP)-tagged rab20 showed a predominantly perinuclear and ER localization pattern and caused wild-type Cx43 to be retained inside the cell. By contrast, mutant EGFP-rab20T19N, which lacks the ability to bind GTP, had no effect on Cx43. These results suggest Cx43 is transported through an intracellular compartment regulated by rab20 along the secretory pathway.
INTRODUCTION
Gap junction proteins (connexins) provide a conduit for intercellular communication by forming channels that enable the diffusion of cytoplasmic ions and small molecules between cells in direct contact (Goldberg et al. Citation2004; Harris Citation2007; Saez et al. Citation2003). An emerging theme in the study of connexins is that formation of gap junction channels is a regulated process (Koval Citation2006; Laird Citation2006). This implies the existence of connexin interacting proteins that control connexin oligomerization and targeting. In fact several proteins have been identified that interact with connexin43 (Cx43), including tubulin, zonula occludens (ZO)-1, ZO-2, connexin interacting protein 85 (CIP85), caveolin-1, and several kinases (Giepmans Citation2006; Moreno and Lau Citation2007; Solan and Lampe Citation2005). Although several connexin interacting proteins were deduced by recognizing motifs on the C-terminal domain, many of these interacting proteins were identified using a yeast two hybrid screen, with the C-terminal domain of Cx43 as “bait” (Giepmans and Moolenaar Citation1998; Jin et al. Citation2000; Lan et al. Citation2005). Yeast two-hybrid screening is clearly a powerful approach, however, it is also subject to limitations. For instance, it is not sensitive to the context of protein expression in mammalian cells. Also, it does not enable the identification of proteins that might regulate connexin trafficking through an indirect interaction.
With this in mind, we developed an alternative screen to identify proteins and pharmacologic agents with the capacity to influence connexin trafficking. As a platform for this assay, we used HeLa cells that were stably transfected with a Cx43 construct containing an endoplasmic reticulum (ER) retention/retrieval signal (HKKSL) (Das Sarma et al. Citation2002, Citation2005; Maza et al. Citation2003, Citation2005). Most of these transfectants retained Cx43-HKKSL in the ER, as demonstrated by immunofluorescence microscopy (see ). However, some overexpressor clones were also identified where Cx43-HKKSL was retained in the perinuclear region of the cell (Das Sarma et al. Citation2005). Previously, we had found that at high levels of expression, Cx43-HKKSL oligomerized and escaped from the ER to localize to the Golgi apparatus (Das Sarma et al. Citation2005). This suggested that the quality control apparatus that regulates connexin oligomerization was saturable and that we might be able to use these cells as a platform to identify factors that potentially regulate Cx43 oligomerization and/or trafficking.
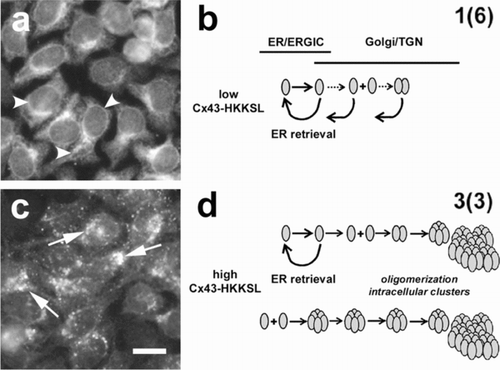
Figure 1 Overexpressed Cx43-HKKSL is retained in the Golgi apparatus. HeLa cells stably transfected with Cx43-HKKSL at either low (a) or high (c) levels show differences in the pattern of intracellular localization by immunofluorescence microscopy. At low levels of expression, the HKKSL tag effectively functions to retain Cx43-HKKSL in the ER (a, arrowheads) and any Cx43-HKKSL which escapes into the Golgi apparatus is retrieved by the di-lysine tag (b). However, when overexpressed, Cx43-HKKSL remains outside the ER and accumulates in the perinuclear region of the cell (c, arrows) where it oligomerizes and forms large intracellular complexes that are resistant to retrieval (d) (Das Sarma et al. Citation2005). Alternatively, high levels of Cx43-HKKSL may force early oligomerization, which, in turn, promotes efflux into the Golgi apparatus and subsequent complex formation. Bar = 10 microns.
In this study we show the utility and limitations of this approach. As starting material, we used a cDNA expression library derived from human kidney cells and created pools of clones that were transfected into a HeLa/Cx43-HHKSL overexpressor cell line 3(3). Transfected cells were screened by immunofluorescence microscopy to identify factors that compensate for overexpression of Cx43-HKKSL by 3(3) cells and promote localization of Cx43-HKKSL to the ER. Pools containing constructs with high activity were serially subdivided until we identified individual cDNAs with activity. Using this approach, we identified a small molecular weight GTPase, rab20, as a protein that regulates Cx43 trafficking by restricting transport from the ER to the Golgi apparatus.
METHODS
Cells and Antibodies
Stably transfected 1(6) and 3(3) HeLa/Cx43-HKKSL cell lines were produced and characterized as previously described (Das Sarma et al. Citation2002, 2005; Maza et al. Citation2003, Citation2005). Rabbit anti-Cx43 was from Sigma (St. Louis, MO), anti-EGFP (enhanced green florescent protein) was from Invitrogen (Carlsbad, CA). Fluorescent and horseradish peroxidase-conjugated secondary antibodies were from Jackson Immunoresearch (West Grove, PA). Triton X-100 was from Roche Molecular Biochemicals (Indianapolis, IN). Tissue culture reagents were from Invitrogen (Carlsbad, CA). Unless otherwise specified, all other reagents were from Sigma.
Immunohistochemistry
For immunofluorescence, cells plated on glass cover slips were fixed and permeabilized with methanol (MeOH)/acetone (1:1), then washed 3× with phosphate-buffered saline (PBS), followed by PBS + 0.5% Triton X-100 and PBS + 0.5% Triton X-100 + 2% goat serum (PBS/GS). The cells were incubated with primary antisera diluted into PBS/GS for 1 h, washed, and then labeled with secondary antisera diluted into PBS/GS. The cells were then washed with PBS, mounted into MOWIOL, visualized by fluorescence microscopy using an Olympus IX-70 microscope system, and imaged with a Hammatzu Orca-1 CCD camera and Image Pro image analysis software (Media Cybernetics, Silver Spring, MD). For screening, we used a 20× UPlanApo oil objective (0.80 NA) and for high resolution images we used a 60× UPlanFl oil objective (0.17 NA).
cDNA Library and Screening
We used a ClonCapture Human Kidney cDNA library in pEXP1 as starting material (Clontech, Palo Alto, CA). The library contained 107 independent clones with an average insert size of 1.5 kb (range 0.5 to 5.0 kb). For amplification 20 × 150 mm LB/amp dishes were plated at 30,000 colony-forming units (CFU)/plate and incubated overnight at 37°C to create 20 pools of clones. For each plate, the colonies were scraped into 5 ml LB/glycerol and split into 1-ml aliquots and frozen at −70°C. To prepare cDNA for transfection, 1 ml of the amplified library was grown overnight in LB/amp liquid culture and the DNA isolated and purified by MaxiPrep (Qiagen). The pools of cDNAs were transiently transfected into HeLa/Cx43-HKKSL 3(3) cells on glass coverslips using Lipofectamine (Invitrogen). Two days after transfection, the cells were fixed, permeabilized, immunostained for using rabbit anti-Cx43 (Sigma) and rhodamine anti-rabbit immunoglobulin G (IgG) (Jackson ImmunoResearch) and screened by immunofluorescence microscopy to manually identify regions where 3(3) cells showed an ER localization pattern. Slides transfected with a high activity pool of cDNAs were identified based on the number of transfected 3(3) cells showing ER-localized Cx43-HKKSL. Depending on the stage of amplification, the pool of clones containing the highest ability to compensate for Cx43-HKKSL overexpression was further subdivided into 20 or 30 equal fractions and analyzed for two more cycles of cDNA preparation and transfection (see ). Once a pool of ∼ 75+ clones was identified as containing cDNAs that could compensate for Cx43-HKKSL overexpression, individual colonies were isolated on LB/AMP plates, picked, and amplified in liquid culture for cDNA isolation by Qiagen MidiPrep. cDNA isolated from individual colonies showing activity were sequenced, using sequences derived from pEXP1 as primers (5′ primer (848–876): 5′-CTGGCTTATC GAAATTAATA CGACTCACT-3′; 3′ primer (1004–977): 5′-TTGGCCGCCC TAGATGCATG CTCGACCT-3′). Clones that encoded for a complete ORF were identified by a BLAST search of the NLM database.
TABLE 1 Summary of cDNA screening
DNA Constructs
myc-tagged connexin43 was produced as described (Das Sarma et al. Citation2002). rab20 cDNA was amplified from the library using High Fidelity DNA polymerase (Roche Diagnostics) with 5′-CCCAAGCTTA CGGGAAGATG AGGAAG CCC-3′ and 5′-GCTCTAGACC TCGAAAGTCA GGCACAAC-3′ as primers. The PCR product was digested with HindIII and XbaI and ligated into pEGFP-C1 and amplified using standard techniques. Point mutated EGFP-rab20T19N was produced using the Qiagen QuikChange mutagenesis kit. DNA for transfection was purified from bacteria using the Qiagen Miniprep kit according to the manufacturer's instructions. All constructs were verified by DNA sequencing prior to use.
RESULTS
At moderate levels of expression, Cx43-HKKSL predominantly localizes to the ER because it has an ER retention/retrieval signal (, ). However, overexpression of Cx43-HKKSL causes it to stably localize to the perinuclear region of the cell (, ). We previously identified this compartment as an aspect of the Golgi apparatus (Das Sarma et al. Citation2005). Also, we previously found that the oligomerization state of Cx43-HKKSL is sensitive to the level of expression (Das Sarma et al. Citation2005). At low to moderate levels of expression, Cx43-HKKSL remained monomeric, whereas, overexpression forced Cx43-HKKSL oligomerization. This suggested that Cx43-HKKSL overexpression in cells such as 3(3) cells saturated the HeLa cell quality control apparatus required to stabilize monomeric Cx43-HKKSL in the ER. This also suggests the possibility that one or more limiting factors required to stabilize Cx43-HKKSL could be added that could compensate for Cx43-HKKSL overexpression by 3(3) cells and thus reflect agents that regulate Cx43 oligomerization and/or trafficking.
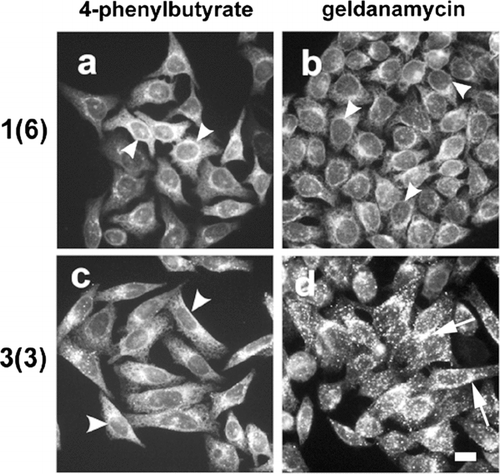
Given this, we developed a screening technique based on fluorescence microscopy to identify these factors. The basic idea behind the screen is that manipulations of 3(3) cells that induce a morphologic changes from a perinuclear to ER localization pattern for Cx43-HKKSL could be readily identified by immunofluorescence microscopy. As an application of this approach, we tested two pharmacologic agents known to affect heat shock proteins for the ability compensate for Cx43-HKKSL overexpression, 4-phenylbutyrate (4-PBA) (Wright et al. Citation2004) and geldanamycin (Hadden et al. Citation2006). 3(3) cells treated overnight with 4-PBA showed an intracellular distribution of Cx43-HKKSL that closely matched 1(6) cells (). However, geldanamycin had little effect on either 1(6) or 3(3) cells. This demonstrated the feasibility and specificity of using 3(3) cells to identify factors that could influence Cx43 trafficking and assembly. Moreover, this is consistent with reports that 4-PBA treatment restores the proper trafficking of the P558S mutant of Cx50 with otherwise misfolds (Berthoud et al. Citation2003). However, 4-PBA, like most drugs, can simultaneously effect several elements of cell behavior. Experiments are currently underway to define a mechanistic basis for the effect of 4-PBA on connexins.
Figure 2 4-Phenylbutyrate restores ER localization of Cx43-HKKSL in 3(3) cells. HeLa cell transfectants stably expressing low (a, b; 1(6)) or high levels (c, d; 3(3)) of Cx43-HKKSL were treated for 18 h with 5 mM 4-PBA (a, c) or 0.1 μ g/ml geldanamycin (b, d), then fixed, permeabilized, and immunostained for Cx43-HKKSL. Following 4-PBA treatment, Cx43-HKKSL expressed by 3(3) cells reverted a distribution similar to 1(6) cells, showing enhanced ER localization (arrowheads). In contrast, geldanamycin had little, if any, effect on either 1(6) or 3(3) cells. Arrows show perinuclear localization of Cx43-HKKSL in 3(3) cells (d). Herbimycin A (3 μ g/ml) also had no obvious effect on either cell type (not shown). Bar = 10 microns.
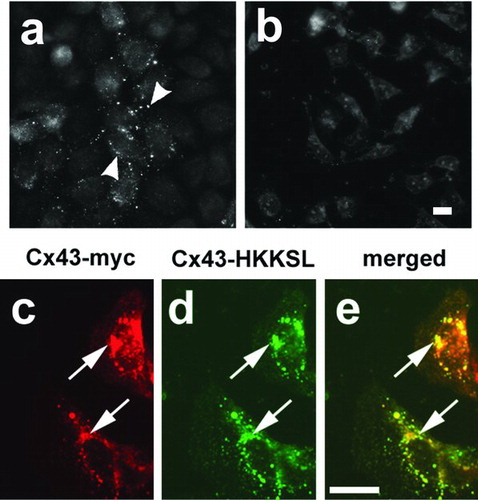
Given the proof of principle that the screening approach was feasible, we used 3(3) cells as a platform to screen a cDNA library produced from human kidney cells. Initially, we were concerned that Cx43 present in the library might give an aberrant false positive, perhaps by facilitating ER localization of Cx43-HKKSL, and thus potentially interfere with the screen. As shown in , the cDNA library did, in fact, contain cDNAs encoding for Cx43. However, added expression of Cx43 alone did not have the capacity to compensate for Cx43-HKKSL overexpression, since 3(3) cells transfected with myc-tagged Cx43 continued to retain Cx43-HKKSL in the perinuclear region of the cells (, , ). Although this is subject to the caveat that we tested this effect using myc-tagged Cx43, Cx43-HKKSL has also been shown to bind and restrict the transport of untagged Cx46 (Das Sarma et al. Citation2002).
Figure 3 Cx43 did not compensate for Cx43-HKKSL overexpression. (a, b) Pools of cDNAs were screened for the presence of Cx43 cDNAs by transfection into HeLa cells, followed by immunofluorescence microscopy. Shown are two pools of clones from round 1 of screening, one of which contained Cx43 cDNA (a; pool 11), another that did not (b; pool 20). Arrowheads show Cx43 localized to gap junctions in (a). 3(3) cells transfected with myc-tagged Cx43 (Cx43-myc) did not show a change in Cx43-HKKSL localization (arrows), suggesting that Cx43 did not compensate for Cx43-HKKSL overexpression by 3(3) cells.
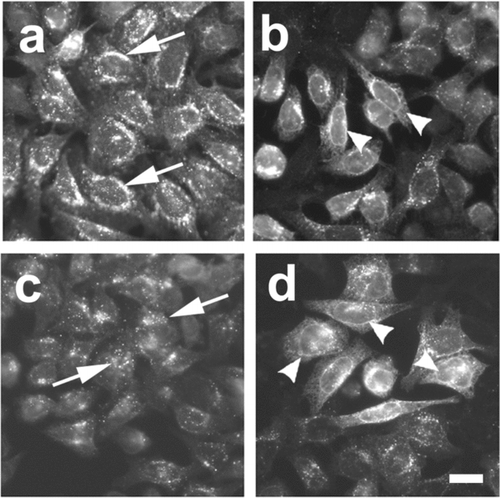
DNA isolated from each pool of colonies was tested in transiently transfected 3(3) cells by immunofluorescence screening. Shown in are examples of fields obtained from pools showing low (, ) and high activity (, ). In each round of enrichment, the pool of clones showing the highest levels of activity was identified, further amplified, subdivided and re-screened. Results from the screen are summarized in . After three cycles of amplification and enrichment, we obtained a pool containing roughly 75 unique cDNAs that showed the ability to compensate for Cx43-HKKSL overexpression. At this point, 81 individual clones were produced and screened.
Figure 4 Assay of cDNA pools that compensate for Cx43-HKKSL overespression by 3(3) cells. Shown are examples of pools of clones which had high activity from round 1 (pool 20) containing ∼ 30,000 unique cDNA clones (a, b) and round 2 (pool 20-7) containing ∼ 1500 unique cDNA clones (c, d). (a and c) Fields of low activity where Cx43-HKKSL remained perinuclear (arrows); (b and d) areas of high activity where several 3(3) cells showed nuclear envelope localization of Cx43-HKKSL (arrowheads). Bar = 10 micron.
Ten individual cDNA clones were identified with the ability to compensate for Cx43-HKKSL overexpression by 3(3) cells. Of these, six were identified to code for expressed human genes (). Three of the remaining clones did not yield interpretable sequences and one corresponded to an intron in the protein kinase theta gene, suggesting a possible false positive. The positive hits in the screen corresponded to a diverse group of proteins ranging from a regulator of mRNA processing (FRG1) to hypothetical proteins of unknown function.
TABLE 2 Proteins identified in the screen
One clone was determined to be a rab-family small-molecular-weight GTPase, rab20 (Lutcke et al. Citation1994). Because we were primarily interested in understanding the regulation of Cx43 trafficking, this protein was further explored. To test whether rab20 has an effect on Cx43 trafficking, an N-terminal EGFP tagged version of the protein was produced, analogous to the approach used to study the function of other rabs (Bucci et al. Citation2000; Handley et al. Citation2007). As shown in , EGFP-rab20 localized predominantly to the perinuclear region of the cell, with low levels of nuclear envelope labeling. This is consistent with the localization of endogenous rab20 in other cell models, where rab20 localizes to the Golgi apparatus (Amillet et al. Citation2006). By sodium dedecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot, EGFP-rab20, had an apparent molecular weight (Mr) close to the predicted size for the fusion protein (∼ 48 kDa).
Figure 5 EGFP-tagged rab20 localizes predominantly to the perinuclear region of the cell. (a, b) Live HeLa cells transfected with EGFP-rab20 (a) or EGFP-rab20T44N (b) were analyzed by fluorescence microscopy. Most EGFP-rab20 localized to the perinuclear region of the cell (arrow), although there also was some nuclear envelope labeling (arrowhead), suggesting possible ER localization. EGFP-rab20T44N had a more uniform appearance, with low levels of apparent nuclear envelope labeling (arrowhead). Bar = 10 micron. (c) Immunoblot analysis of untransfected HeLa cells (un), cells transfected with EGFP-rab20 (wt) or EGFP-rab20T19N (T19N) using anti-EGFP for detection. Transfected cells show a specific band of near the predicted mass for the fusion proteins of ∼ 48 kDa.
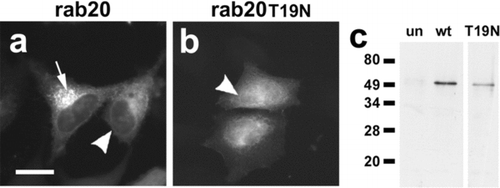
In addition to a tagged version of wild-type rab20, we also produced a T19N mutant in the conserved sequence VGKSTL (EGFP-rab20T19N), which is predicted to be inactive and unable to bind GTP, based on the T22N rab7 mutant (Bucci et al. Citation2000). In contrast to EGFP-rab20, EGFP-rab20T19N transfected into HeLa cells had a more uniform distribution and filled the nucleus, although there were some hints of nuclear envelope labeling ().
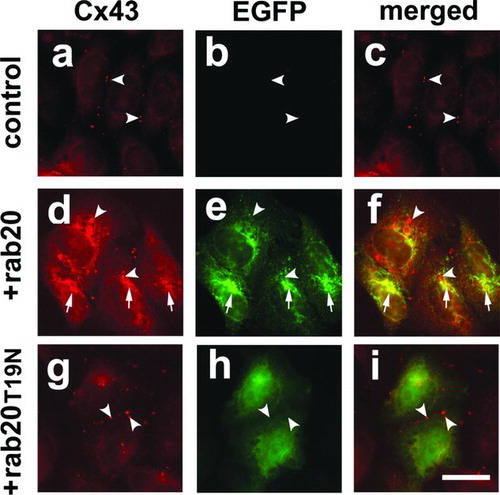
To determine whether rab20 had an effect on Cx43, we transfected EGFP-rab20 or EGFP-rab20T19N into HeLa/Cx43 cells. EGFP-rab20 had a profound effect on Cx43 and inhibited trafficking to the plasma membrane (, , ). Instead, Cx43 was retained within the cell and showed considerable co-localization with EGFP-rab20. In contrast, for HeLa/Cx43 cells transfected with EGFP-rab20T19N, Cx43 was mostly localized to gap junctions and appeared similar to untransfected controls. Much of the intracellular Cx43 labeling in HeLa/Cx43 cells transfected with EGFP-rab20 suggested ER retention of Cx43, because there was prominent nuclear envelope labeling. ER retention of Cx43 in cells expressing EGFP-rab20 is consistent with the results from the screen, where rab20 enhanced the ER retention of Cx43-HKKSL expressed by 3(3) cells. However, there also were some vesicles filled with Cx43 surrounded by a ring of EGFP-rab20 labeling. Taken together, these results support the notion that rab20 has the ability to regulate the trafficking of Cx43 along the secretory pathway.
Figure 6 EGFP-rab20 inhibits Cx43 trafficking to the plasma membrane. Shown is immunofluorescence analysis of HeLa/Cx43 cells (a–c), HeLa/Cx43 cells transfected with EGFP-rab20 (d–f) or EGFP-rab20T19N (g–i). HeLa/Cx43 cells show Cx43 localized to gap junctions (arrowheads). Cells expressing EGFP-rab20 had partial co-localization with Cx43 in the perinuclear region of the cell (arrows), although there also were intracellular vesicles labeled for Cx43 that did contain EGFP-rab20 (arrowheads). By contrast, cells expressing EGFP-rab20T19N did not show this effect on Cx43, which assembled into gap junctions (arrowheads). Bar = 10 microns.
DISCUSSION
In our screen for factors that control connexin trafficking and assembly, we identified rab20 as a protein with the ability to regulate Cx43 transport. rab family proteins are small-molecular-weight GTPases, which associate with distinct intracellular membrane compartments and regulate membrane traffic (Zerial and McBride Citation2001). The best characterized rab function is regulation of membrane fusion by cycling between an active, membrane associated, GTP-bound state and an inactive, GDP-bound state (Barbieri et al. Citation1998; Cao and Barlowe Citation2000; Grindstaff et al. Citation1998). Once at the membrane, rabs orchestrate the assembly of membrane fusion machinery (Carr et al. Citation1999). Roles for rabs in vesicle budding and motility have also been described (Echard et al. Citation1998; Riederer et al. Citation1994). In addition, rabs define organelle subdomains by forming distinct clusters in different membrane regions (Sheff et al. Citation1999; Sonnichsen et al. Citation2000; Trischler et al. Citation1999). With respect to Cx43 trafficking, a rab-defined subdomain would be of interest, because it might define a compartment where Cx43 and related connexins are oligomerized.
Soluble protein cofactors, known as effectors, help regulate rab activity by performing functions such as promoting GDP-GTP exchange (GDP exchange factors [GEFs]) or activating rab GTPase activity (GTPase activiating proteins [GAPs]). Recently, a connexin interacting protein, CIP85, was identified that binds to Cx43 and shares homology with rab-GAP proteins (Lan et al. Citation2005). CIP85 binds directly to a PLSP domain in the cytoplasmic tail of Cx43 and colocalizes with Cx43 at the plasma membrane. There is also a pool of intracellular CIP85 and CIP85 overexpression enhanced Cx43 turnover, suggesting a role in regulating Cx43 trafficking. Although a direct interaction of CIP85 with rabs has not yet been demonstrated, our results are consistent with the possibility that CIP85 or a related protein might mediate an interaction between rab20 and Cx43. Whether rab20 helps regulate Cx43 trafficking through a direct interaction with Cx43, an interaction with Cx43 as part of a multiprotein complex, or indirectly by altering a general membrane trafficking pathway remains to be determined.
rab-mediated signaling has been implicated in several human diseases (Seabra et al. Citation2002). Modulation of rab expression also could provide a therapeutic benefit for other classes of human disease. For instance, manipulating rab7 and rab9 expression has been proposed as a possible treatment for sphingolipid storage diseases (Choudhury et al. Citation2002). To date, the function of rab20 has not been defined, although our data suggest a role for rab20 in regulating membrane transport from the ER to the Golgi apparatus. Because rab20 inhibited transport of Cx43 along the secretory pathway, the ability of rab20 to compensate for Cx43-HKKSL overexpression was likely due to a comparable effect, where rab20 inhibited Cx43-HKKSL efflux from the ER.
rab20 is highly expressed in a number of epithelia, including kidney, intestine, and lung (Lutcke et al. Citation1994; McMurtrie et al. Citation1997). Intriguingly, increased rab20 expression was found in 12 out of 21 patients with a class of preleukemic disorders known as myelodysplastic syndromes (Pellagatti et al. Citation2004) and in 15 of 18 exocrine pancreatic adenocarcinomas (Amillet et al. Citation2006), suggesting a role for rab20 in tumor progression. The ability of rab20 to interfere with formation of gap junctions by Cx43 fits well with the association of decreased Cx43 activity with cancer (Goldberg et al. Citation2000; Shao et al. Citation2005). Further work is required to determine whether this is the case.
We also identified 4-PBA as a pharmacologic agent with the ability to compensate for Cx43-HKKSL overexpression. 4-PBA restores the proper trafficking of several classes of misfolded proteins, including mutant Cx50 (Berthoud et al. Citation2003), CFTR-Δ F508 (Rubenstein et al. Citation1997), p-Z-α 1-anti-trypsin (Burrows et al. Citation2000) and mutant surfactant protein C (Wang et al. Citation2003). Given this, we tested whether 4-PBA could compensate for Cx43-HKKSL overexpression by 3(3). As shown in , this was the case. The cell response to 4-PBA is complex and includes changes in gene expression and inhibition of histone acetylation (Chen et al. Citation1997; Wright et al. Citation2004), which can also affect Cx43 expression (Ammerpohl et al. Citation2004; Asklund et al. Citation2004; Hattori et al. Citation2007; Khan et al. Citation2007). Interestingly, geldanamycin, an agent also shown to alter Cx43 regulation (Carystinos et al. Citation2003; Lidington et al. Citation2000, Citation2002; Rodriguez-Sinovas et al. Citation2006) had no effect on 3(3) cells.
One effect of 4-PBA is to increase heat shock protein 70 (Hsp70) relative to Hsc70 expression (Rubenstein and Lyons Citation2001; Rubenstein and Zeitlin Citation2000). However, we found that overexpression of tetracycline-inducible isoforms of either Hsp70 or heat shock cognate protein 70 (Hsc70) transfected into 3(3) cells was not sufficient to compensate for Cx43-HKKSL overexpression (not shown). Although this does not rule out a role for these proteins in regulating connexin folding/oligomerization, the lack of activity by these heat shock proteins suggested that the effect of 4-PBA on 3(3) cells was due to some other factor. Current work is underway to identify factors upregulated by 4-PBA with the potential to regulate connexin trafficking or oligomerization.
We used HKKSL-tagged Cx43 as the basis for our screen for several reasons. First, Cx43-HKKSL overexpressors, such as 3(3) cells, that had a distinct perinuclear pattern of Cx43-HKKSL localization were readily distinguishable from cells where Cx43-HKKSL was ER localized, making manual screening possible by immunofluorescence. Also, because compensation for Cx43-HKKSL overexpression leads to ER retention of this protein, the assay did not require that cells properly interact with neighboring cells, as would be the case for an assay which requires gap junction formation. On the other hand, because Cx43-HKKSL does not reflect the normal itinerary and processing of Cx43, screens based on Cx43-HKKSL run the risk of identifying proteins that do not regulate wild type Cx43. For instance, a class of potential false positives are elements of the coatomer protein complex, because overexpression of the HKKSL tag might saturate the ER retention/retrieval pathway which requires these proteins (Andersson et al. Citation1999).
Another rationale for using a Cx43-HKKSL overexpressors as a screening platform was that we anticipated it might favor proteins that directly interact with Cx43-HKKSL to reverse oligomerization (see ). Although the diverse nature of positive hits suggests that this is may not be the case, the proteins of unknown function that were identified in the screen may directly interact with Cx43 (). By contrast, several of the candidate proteins identified in the screen are more likely to regulate connexins through changes in cell signaling or differentiated state. For instance, although decorin B has not been shown to regulate intercellular communication, cross-talk between integrins, other extracellular matrix proteins, and connexins has previously been demonstrated (Isakson et al. Citation2001; Kalra et al. Citation2006; Lampe et al. Citation1998). A similar case could be made for FRG1, which is associated with an autosomal dominant disease, facioscapulohumeral muscular dystrophy (Gabellini et al. Citation2006).
Several mutant connexins associated with human disease show defects in intracellular trafficking (Berthoud et al. Citation2003; Gong et al. Citation2006; McLachlan et al. Citation2005; VanSlyke et al. Citation2000). Thus, a screen comparable to the one described here but based on wild-type or mutant connexins could potentially help identify factors that influence intercellular communication and/or compensate for mutant connexins, regardless of whether these factors mediate a direct or indirect interaction with a connexin.
Here, cells were manually processed and analyzed by immunofluorescence which was admittedly tedious. However, progress continues to be made in automating morphologic screens based on fluorescence microscopy and pattern recognition (Glory and Murphy Citation2007). In particular, disease-related mutant connexins that show intracellular retention may be amenable to an automated morphologic screen, as a high throughput screen for agents with the ability to restore proper connexin trafficking to the plasma membrane. Sensitivity could be increased by using fluorescent markers for intracellular compartments (Giepmans et al. Citation2006) and fluorescently tagged connexins (Laird et al. Citation2001), which would enable analysis of live cells. The success of such a screen will require the ability to distinguish between intracellular puncta and gap junction–localized connexins and whether restoration of connexin transport to the plasma membrane is sufficient to reverse the disease phenotype.
The authors thank Dr. Crislyn D'Souza-Schorey for advice related to rab constructs. This work was supported by National Institutes of Health grants R01-GM061012 and R01-HL083120 (MK).
REFERENCES
- Amillet J M, Ferbus D, Real F X, Antony C, Muleris M, Gress T M, Goubin G. Characterization of human Rab20 overexpressed in exocrine pancreatic carcinoma. Hum Pathol 2006; 37: 256–263
- Ammerpohl O, Thormeyer D, Khan Z, Appelskog I B, Gojkovic Z, Almqvist P M, Ekstrom T J. HDACi phenylbutyrate increases bystander killing of HSV-tk transfected glioma cells. Biochem Biophys Res Commun 2004; 324: 8–14
- Andersson H, Kappeler F, Hauri H P. Protein targeting to endoplasmic reticulum by dilysine signals involves direct retention in addition to retrieval. J Biol Chem 1999; 274: 15080–15084
- Asklund T, Appelskog I B, Ammerpohl O, Ekstrom T J, Almqvist P M. Histone deacetylase inhibitor 4-phenylbutyrate modulates glial fibrillary acidic protein and connexin 43 expression, and enhances gap-junction communication, in human glioblastoma cells. Eur J Cancer 2004; 40: 1073–1081
- Barbieri M A, Hoffenberg S, Roberts R, Mukhopadhyay A, Pomrehn A, Dickey B F, Stahl P D. Evidence for a symmetrical requirement for Rab5-GTP in in vitro endosome-endosome fusion. J Biol Chem 1998; 273: 25850–25855
- Berthoud V M, Minogue P J, Guo J, Williamson E K, Xu X, Ebihara L, Beyer E C. Loss of function and impaired degradation of a cataract-associated mutant connexin50. Eur J Cell Biol 2003; 82: 209–221
- Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: A key to lysosome biogenesis. Mol Biol Cell 2000; 11: 467–480
- Burrows J A, Willis L K, Perlmutter D H. Chemical chaperones mediate increased secretion of mutant alpha 1-antitrypsin (alpha 1-AT) Z: A potential pharmacological strategy for prevention of liver injury and emphysema in alpha 1-AT deficiency. Proc Natl Acad Sci U S A 2000; 97: 1796–1801
- Cao X, Barlowe C. Asymmetric requirements for a Rab GTPase and SNARE proteins in fusion of COPII vesicles with acceptor membranes. J Cell Biol 2000; 149: 55–66
- Carr C M, Grote E, Munson M, Hughson F M, Novick P J. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J Cell Biol 1999; 146: 333–44
- Carystinos G D, Kandouz M, Alaoui-Jamali M A, Batist G. Unexpected induction of the human connexin 43 promoter by the ras signaling pathway is mediated by a novel putative promoter sequence. Mol Pharmacol 2003; 63: 821–831
- Chen W Y, Bailey E C, McCune S L, Dong J Y, Townes T M. Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylase. Proc Natl Acad Sci U S A 1997; 94: 5798–803
- Choudhury A, Dominguez M, Puri V, Sharma D K, Narita K, Wheatley C L, Marks D L, Pagano R E. Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J Clin Invest 2002; 109: 1541–50
- Das Sarma J, Das S, Koval M. Regulation of connexin43 oligomerization is saturable. Cell Commun Adhes 2005; 12: 237–47
- Das Sarma J, Wang F, Koval M. Targeted gap junction protein constructs reveal connexin-specific differences in oligomerization. J Biol Chem 2002; 277: 20911–20918
- Echard A, Jollivet F, Martinez O, Lacapere J J, Rousselet A, Janoueix-Lerosey I., Goud B. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science 1998; 279: 580–585
- Gabellini D, D'Antona G., Moggio M, Prelle A, Zecca C, Adami R, Angeletti B, Ciscato P, Pellegrino M A, Bottinelli R, Green M R, Tupler R. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature 2006; 439: 973–977
- Giepmans B N. Role of connexin43-interacting proteins at gap junctions. Adv Cardiol 2006; 42: 41–56
- Giepmans B N, Adams S R, Ellisman M H, Tsien R Y. The fluorescent toolbox for assessing protein location and function. Science 2006; 312: 217–224
- Giepmans B N, Moolenaar W H. The gap junction protein connexin43 interacts with the second PDZ domain of the zona occludens-1 protein. Curr Biol 1998; 8: 931–934
- Glory E, Murphy R F. Automated subcellular location determination and high-throughput microscopy. Dev Cell 2007; 12: 7–16
- Goldberg G S, Bechberger J F, Tajima Y, Merritt M, Omori Y, Gawinowicz M A, Narayanan R, Tan Y, Sanai Y, Yamasaki H, Naus C C, Tsuda H, Nicholson B J. Connexin43 suppresses MFG-E8 while inducing contact growth inhibition of glioma cells. Cancer Res 2000; 60: 6018–6026
- Goldberg G S, Valiunas V, Brink P R. Selective permeability of gap junction channels. Biochim Biophys Acta 2004; 1662: 96–101
- Gong X Q, Shao Q, Lounsbury C S, Bai D, Laird D W. Functional characterization of a GJA1 frame-shift mutation causing oculodentodigital dysplasia and palmoplantar keratoderma. J Biol Chem 2006; 281: 31801–31811
- Grindstaff K K, Yeaman C, Anandasabapathy N, Hsu S C, Rodriguez-Boulan E, Scheller R H, Nelson W J. Sec6/8 complex is recruited to cell-cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell 1998; 93: 731–740
- Hadden M K, Lubbers D J, Blagg B S. Geldanamycin, radicicol, and chimeric inhibitors of the Hsp90 N-terminal ATP binding site. Curr Top Med Chem 2006; 6: 1173–1182
- Handley M T, Haynes L P, Burgoyne R D. Differential dynamics of Rab3A and Rab27A on secretory granules. J Cell Sci 2007; 120: 973–984
- Harris A L. Connexin channel permeability to cytoplasmic molecules. Prog Biophys Mol Biol 2007; 94: 120–143
- Hattori Y, Fukushima M, Maitani Y. Non-viral delivery of the connexin 43 gene with histone deacetylase inhibitor to human nasopharyngeal tumor cells enhances gene expression and inhibits in vivo tumor growth. Int J Oncol 2007; 30: 1427–1439
- Isakson B E, Lubman R L, Seedorf G J, Boitano S. Modulation of pulmonary alveolar type II cell phenotype and communication by extracellular matrix and KGF. Am J Physiol Cell Physiol 2001; 281: C1291–C1299
- Jin C, Lau A F, Martyn K D. Identification of connexin-interacting proteins: Application of the yeast two-hybrid screen. Methods 2000; 20: 219–231
- Kalra J, Shao Q, Qin H, Thomas T, Alaoui-Jamali M A, Laird D W. Cx26 inhibits breast MDA-MB-435 cell tumorigenic properties by a gap junctional intercellular communication-independent mechanism. Carcinogenesis 2006; 27: 2528–2537
- Khan Z, Akhtar M, Asklund T, Juliusson B, Almqvist P M, Ekstrom T J. HDAC inhibition amplifies gap junction communication in neural progenitors: Potential for cell-mediated enzyme prodrug therapy. Exp Cell Res 2007; 313: 2958–2967
- Koval M. Pathways and control of connexin oligomerization. Trends Cell Biol 2006; 16: 159–166
- Laird D W. Life cycle of connexins in health and disease. Biochem J 2006; 394: 527–543
- Laird D W, Jordan K, Shao Q. Expression and imaging of connexin-GFP chimeras in live mammalian cells. Methods Mol Biol 2001; 154: 135–142
- Lampe P D, Nguyen B P, Gil S, Usui M, Olerud J, Takada Y, Carter W G. Cellular interaction of integrin alpha3beta1 with laminin 5 promotes gap junctional communication. J Cell Biol 1998; 143: 1735–1747
- Lan Z, Kurata W E, Martyn K D, Jin C, Lau A F. Novel rab GAP-like protein, CIP85, interacts with connexin43 and induces its degradation. Biochemistry 2005; 44: 2385–2396
- Lidington D, Ouellette Y, Tyml K. Endotoxin increases intercellular resistance in microvascular endothelial cells by a tyrosine kinase pathway. J Cell Physiol 2000; 185: 117–125
- Lidington D, Tyml K, Ouellette Y. Lipopolysaccharide-induced reductions in cellular coupling correlate with tyrosine phosphorylation of connexin 43. J Cell Physiol 2002; 193: 373–379
- Lutcke A, Parton R G, Murphy C, Olkkonen V M, Dupree P, Valencia A, Simons K, Zerial M. Cloning and subcellular localization of novel rab proteins reveals polarized and cell type-specific expression. J Cell Sci 1994; 107: 3437–3448, (Pt 12)
- Maza J, Mateescu M, Sarma J D, Koval M. Differential oligomerization of endoplasmic reticulum-retained connexin43/connexin32 chimeras. Cell Commun Adhes 2003; 10: 319–322
- Maza J, Das Sarma J, Koval M. Defining a minimal motif required to prevent connexin oligomerization in the endoplasmic reticulum. J Biol Chem 2005; 280: 21115–21121
- McLachlan E, Manias J L, Gong X Q, Lounsbury C S, Shao Q, Bernier S M, Bai D, Laird D W. Functional characterization of oculodentodigital dysplasia-associated Cx43 mutants. Cell Commun Adhes 2005; 12: 279–292
- McMurtrie E B, Barbosa M, Zerial M, Kingsmore S F. Genetic mapping of Rab20 on mouse chromosome 8. Mammal Genome 1997; 8: 291–292
- Moreno A P, Lau A F. Gap junction channel gating modulated through protein phosphorylation. Prog Biophys Mol Biol 2007; 94: 107–119
- Pellagatti A, Esoof N, Watkins F, Langford C F, Vetrie D, Campbell L J, Fidler C, Cavenagh J D, Eagleton H, Gordon P, Woodcock B, Pushkaran B, Kwan M, Wainscoat J S, Boultwood J. Gene expression profiling in the myelodysplastic syndromes using cDNA microarray technology. Br J Haematol 2004; 125: 576–583
- Riederer M A, Soldati T, Shapiro A D, Lin J, Pfeffer S R. Lysosome biogenesis requires Rab9 function and receptor recycling from endosomes to the trans-Golgi network. J Cell Biol 1994; 125: 573–582
- Rodriguez-Sinovas A, Boengler K, Cabestrero A, Gres P, Morente M, Ruiz-Meana M, Konietzka I, Miro E, Totzeck A, Heusch G, Schulz R, Garcia-Dorado D. Translocation of connexin 43 to the inner mitochondrial membrane of cardiomyocytes through the heat shock protein 90-dependent TOM pathway and its importance for cardioprotection. Circ Res 2006; 99: 93–101
- Rubenstein R C, Egan M E, Zeitlin P L. In vitro pharmacologic restoration of CFTR-mediated chloride transport with sodium 4-phenylbutyrate in cystic fibrosis epithelial cells containing delta F508-CFTR. J Clin Invest 1997; 100: 2457–2465
- Rubenstein R C, Lyons B M. Sodium 4-phenylbutyrate downregulates HSC70 expression by facilitating mRNA degradation. Am J Physiol Lung Cell Mol Physiol 2001; 281: L43–L51
- Rubenstein R C, Zeitlin P L. Sodium 4-phenylbutyrate downregulates Hsc70: Implications for intracellular trafficking of DeltaF508-CFTR. Am J Physiol Cell Physiol 2000; 278: C259–C267
- Saez J C, Berthoud V M, Branes M C, Martinez A D, Beyer E C. Plasma membrane channels formed by connexins: Their regulation and functions. Physiol Rev 2003; 83: 1359–1400
- Seabra M C, Mules E H, Hume A N. Rab GTPases, intracellular traffic and disease. Trends Mol Med 2002; 8: 23–30
- Shao Q, Wang H, McLachlan E., Veitch G I, Laird D W. Down-regulation of Cx43 by retroviral delivery of small interfering RNA promotes an aggressive breast cancer cell phenotype. Cancer Res 2005; 65: 2705–2711
- Sheff D R, Daro E A, Hull M, Mellman I. The receptor recycling pathway contains two distinct populations of early endosomes with different sorting functions. J Cell Biol 1999; 145: 123–139
- Solan J L, Lampe P D. Connexin phosphorylation as a regulatory event linked to gap junction channel assembly. Biochim Biophys Acta 2005; 1711: 154–163
- Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, Rab11. J Cell Biol 2000; 149: 901–914
- Trischler M, Stoorvogel W, Ullrich O. Biochemical analysis of distinct Rab5-and Rab11-positive endosomes along the transferrin pathway. J Cell Sci 1999; 112: 4773–4783, (Pt 24)
- VanSlyke J K, Deschenes S M, Musil L S. Intracellular transport, assembly, and degradation of wild-type and disease-linked mutant gap junction proteins. Mol Biol Cell 2000; 11: 1933–1946
- Wang W J, Mulugeta S, Russo S J, Beers M F. Deletion of exon 4 from human surfactant protein C results in aggresome formation and generation of a dominant negative. J Cell Sci 2003; 116: 683–692
- Wright J M, Zeitlin P L, Cebotaru L, Guggino S E, Guggino W B. Gene expression profile analysis of 4-phenylbutyrate treatment of IB3-1 bronchial epithelial cell line demonstrates a major influence on heat-shock proteins. Physiol Genomics 2004; 16: 204–211
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2001; 2: 107–117
