Abstract
Kru¨ppel like factor 4 (KLF4) is a transcription factor that regulates genes related to differentiation and proliferation. KLF4 also plays a role in metastasis via epithelial to mesenchymal transition. Here, we investigate the function of Klf4 in migration and invasion using mouse embryonic fibroblasts and the RKO human colon cancer cell line. Compared to wild-type, cells lacking Klf4 exhibited increased migration-associated phenotypes. In addition, overexpression of Klf4 in Klf4−/− MEFs attenuated the presence of stress fibers to wild-type levels. An invasion assay suggested that lack of Klf4 resulted in increased invasive capacity. Finally, analysis of RhoA showed elevated RhoA activity in both RKO and MEF cells. Taken together, our results strongly support the novel role of KLF4 in a post-translational regulatory mechanism where KLF4 indirectly modulates the actin cytoskeleton morphology via activity of RhoA in order to inhibit cellular migration and invasion.
Introduction
KLF4 is a zinc-finger transcription factor belonging to the Kru¨ppel-like family that is involved in the regulation of a diverse group of biological processes such as apoptosis, differentiation, and proliferation (Garrett-Sinha et al. Citation1996; Shields et al. Citation1996; Hagos et al. Citation2009). While the function of KLF4 as a tumor suppressor has been demonstrated in multiple forms of cancers, its role as an oncogene has also been proposed due to its upregulation in squamous cell carcinoma and breast cancer, which suggests that its regulatory role in tumorigenesis is context-dependent (Ohnishi et al. Citation2003; Pandya et al. Citation2004; Zhao et al. Citation2004; Huang et al. Citation2005; Wei et al. Citation2005; Rowland and Peeper Citation2006; Zammarchi et al. Citation2010). We have previously shown that Klf4 knockout in MEFs is associated with genomic instability and impaired autophagy during oxidative stress, resulting in increased DNA damage and apoptosis (Hagos et al. Citation2009; Liu et al. Citation2014; Liu et al. Citation2015). The genomic instability associated with lack of Klf4 was additionally found to be reversible by transfecting KLF4-expression plasmids to Klf4−/− MEFs (El-Karim et al. Citation2013). Furthermore, KLF4 prevents epithelial to mesenchymal transition and metastasis of breast cancer both by inhibiting Snail and activating E-cadherin (Yori et al. Citation2011). However, the specifics of the many potential tumor suppressor pathways in which KLF4 is involved are yet to be precisely defined.
Evidence suggests that KLF4 deficiency may be associated with cell migration and adhesion, which are important characteristics of tumor cells (Li et al. Citation2012; Lv et al. Citation2016). The organization of the actin cytoskeleton is closely related to the overall shape and morphology of the cell. In MEFs, more migratory cells have elongated phenotypes, which have been linked to morphological changes and alterations in cell motility (Goulimari et al. Citation2005, Thievessen et al. Citation2015). This raised a question as to whether KLF4 might play a role in the regulation of actin reorganization, a critical step in regulation of cell motility (Pollard and Borisy Citation2003; Yamazaki et al. Citation2005).
With respect to the regulation of cytoskeletal dynamics, proteins in the Rho GTPase family were considered to be attractive candidates, as Rho A, B, and C expression have been extensively shown to modify actin polymerization (Ridley and Hall Citation1992; Raftopoulou and Hall Citation2004). Specifically, RhoA activity is known to induce transformation and motility-associated actin structures, including stress fibers, which can promote metastasis (Ridley Citation2013; Lei et al. Citation2014). The activity of Rho proteins, as in other small GTPases, is controlled via binding to either GTP, to induce an activated conformation, or GDP, for the inactivated state (Boguski and McCormick Citation1993). This process is regulated by Guanine nucleotide exchange factor (GEF), which acts as a catalyst for stimulating the release of GDP in exchange for GTP, and GTPase-activating protein (GAP), which increases the rate of GTPase activity with respect to Rho proteins (Van Aelst and D’Souza-Schorey Citation1997).
In the present study, we sought to investigate the mechanism by which Klf4 inhibits migration and invasion. Here, we show that MEFs and RKO cells lacking Klf4 display increased frequency of morphology and actin reorganization favorable to migration. The morphological variation was supported by increased rates of migration and invasion for cells without Klf4 expression. We further propose that this phenomenon involves a mechanism downstream of Klf4 that affects GAP or GEF proteins with respect to RhoA activity rather than directly affecting RhoA expression at the transcriptional or translational level.
Materials and methods
Cell culture, reagents, and drug treatment
Spontaneously immortalized primary MEFs, either wild-type (Klf4+/+) or null (Klf4−/−) for Klf4, were derived from terminated 13.5-day old embryos of crossbred mice heterozygous for Klf4 (Klf4+/−) on a C57BL/6 background to generate the homozygous knockout and littermate wildtype (Katz et al. Citation2002). Cells were passed about every three days following seeding with the 3T3 protocol (Todaro and Green Citation1963). All experiments were performed post-senescence and cells were discarded past passage 25 to maintain genomic integrity. The RKO (RKO-EcR-KLF4) cell line was derived from a human colon cancer cell line and stably transfected with the pAdLoxEGI-KLF4 plasmid, as previously described (Chen et al. Citation2001). The plasmid contains the ecdysone-inducible promoter (EcRE), which is not naturally expressed by the human cell line, and a full-length KLF4 gene that is naturally expressed at undetectable levels (Dang et al. Citation2001). The EcRE gene is conjugated to an enhanced GFP gene and accompanied by an internal ribosomal entry site. Therefore, KLF4 is conditionally expressed in RKO cells via the addition of Ponasterone-A (PA) (Sigma-Aldrich, St. Louis, MO, #P3490) and EtOH served as the solvent control (Dang et al. Citation2003). Both cell lines were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cells were incubated at 37 °C in a 5% CO2 environment. Cells were grown to appropriate confluency (60–70%) and then treated with 5 µM PA in EtOH for 3 days. For ROCK inhibition, the cells were treated with 10 μM Y-27632 (Sigma-Aldrich, St. Louis, MO, #Y0503). For serum starvation, the cells were washed with PBS and then incubated in 1% FBS.
Western blotting
Cells were grown to appropriate confluency after which protein samples were extracted in lysis buffer, ground up with acid-treated glass beads, and then denatured by heating to 98 °C for 5 minutes. The lysates were loaded onto Mini-PROTEAN® TGX™ Precast Gels (Bio-Rad Laboratories, Hercules, CA, #4561096) in Tri/Glycine/SDS buffer (Bio-Rad Laboratories, Hercules, CA, #1610732) for electrophoresis. The gel was then transferred to a nitrocellulose membrane in Tris/glycine transfer buffer (Bio-Rad Laboratories, Hercules, CA, #1610734) with 10% methanol. The membrane was immunoblotted with primary antibodies, which in this study consisted of: GAPDH (housekeeping control, #97166), RhoA, (Cell Signaling, Danvers, MA, #2117), and RhoC (AbCam, Branford, CT, #ab64659). The HRP-conjugated secondary antibodies used were anti-mouse and anti-rabbit (Cell Signaling, Danvers, MA, #7076, #7074, respectively). The protein expression was then visualized by an Immun-Star™ HRP Chemiluminescence Kit (Bio-Rad Laboratories, Hercules, CA, #1705070) and ChemiDoc™ XRS + System (Bio-Rad Laboratories, Hercules, CA, #1708265) and imaged through Image Lab on a Gel Doc system.
Phalloidin staining and morphology identification
Cells were fixed with 3.7% formaldehyde and then stained with DAPI and 100 nM Acti-stain 555 Phalloidin (Cytoskeleton, Inc., #PHDH1-A) for 30 minutes. Stained cells were visualized on an Olympus U-RFL-T fluorescent microscope and imaged using Infinity Analyze. For phenotypic analysis, actin stress fibers were qualified by the presence of directional non-uniform, filamentous structures limited to the interior of the cell. Filopodia were identified as thin actin structures that protruded beyond the cell body. Lamellipodia were identified as semicircular actin structures on the leading periphery of the cell body (Lehtimaki et al. Citation2017). Cells were plated at low confluency and allowed to incubate under standard cell culture conditions. After treatment conditions, cells were imaged at 10 random locations within the plate using brightfield microscopy on an Olympus U-RFL-T. In each image, the total cell number was counted (n > 300 cells per condition), and each individual cell was manually determined positive or negative for stress fiber, filopodia, and lamellipodia to create percentages. In conditions with plasmid transfection, only GFP positive cells were counted (n > 150). For brightfield morphology analysis, migratory phenotypes were identified based on cell elongation for MEFs and a non-polar shape for RKO cells as supported by primary literature (Goulimari et al. Citation2005; Takahashi et al. Citation2014).
Wound-healing assay
Cells were grown to high confluency (about 90%) and then switched to 1% FBS conditions before being scraped with the tip of a 200 µL pipette adjacent to a reference mark. Brightfield microscopy was used to image initial size of scrape at time 0. Migratory rates were determined by comparing the area at initial and final time using ImageJ calculations. Serum starvation was utilized in order to compensate for the effects of proliferation (Liu et al. Citation2015).
Invasion assay
QCM ECMatrix Cell Invasion Assay kit (EMD Millipore, Darmstadt, Germany; #ECM550) was used to examine the invasive characteristics of the cells. Cells were counted and plated in equal numbers (1 × 106 cells/ml) onto an invasion chamber which consisted of tissue culture plates with cell culture inserts that contain polycarbonate membranes with 8 μM pores layered with thin ECMatrix™. During 72 hours of incubation in the nutrient gradient, invasive cells were allowed to migrate from the 0.1% FBS top layer through the thin ECM layer into the lower 10% FBS supplemented DMEM and cling to the bottom of the membrane. The invasive cells were stained with Crystal Violet and then imaged via brightfield microscopy. The number of stained cells in each well was counted and the average number of cells for each data point was calculated.
Transient transfection
MEF cells were transfected with 5 μg/μL of DNA, either encoding GFP or Klf4 conjugated to GFP on a EGFP plasmid backbone, using the Lipofectamine 3000 Transfection Reagent Protocol (Thermo Fisher, Waltham, MA, #L3000015) at 70% confluency as previously described (Liu et al. Citation2015). The efficiency of the transfection was observed under an Olympus IX51 microscope 24 hours post-transfection.
RhoA activation assay
Cells grown under normal conditions were treated in serum starvation condition (1% FBS) for 6 hours, after which the samples were collected and snap frozen in liquid nitrogen to be preserved. After the protein quantity was made equivalent across samples by measuring the protein concentration using a Bradford reagent via the NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA), Rhotekin-RBD beads and other reagents from the RhoA pulldown activation assay Biochem Kit (Cytoskeleton, Inc., Denver, CO, #BK036-S) were added to the protein lysates and then centrifuged as described in the product protocol provided by the company. A Western Blot was then performed on the proteins that remained bound to the beads using a special RhoA primary antibody provided by the company.
Statistical analysis
GraphPad QuickCalcs software was used to perform an unpaired t test to evaluate the difference between trials where indicated. p Values below .05 (p < .05) were considered statistically significant. Graphpad prism was used to generate some figures. In all figures, error bars indicate standard deviation.
Results and discussion
Klf4-deficient cells exhibit an altered morphology associated with migration when compared to cells expressing Klf4
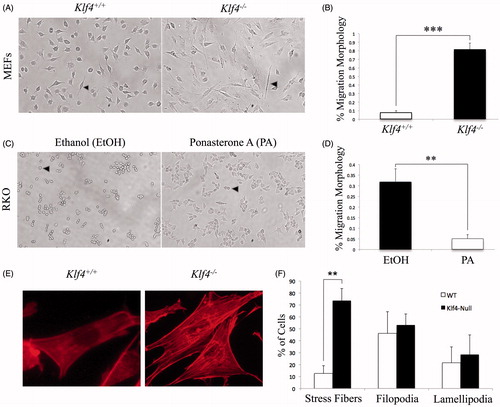
A visual comparison of Klf4+/+ and Klf4−/− MEFs under brightfield revealed consistent differences in morphology. Klf4+/+ cells showed a less protruding morphology typical of fibroblasts while Klf4−/− cells were more elongated (). Quantification of individual cell morphologies between Klf4+/+ and Klf4−/− MEFs showed that the difference was statistically significant at a p value of <.0001 (). Furthermore, our data are consistent with previous research that found an association of elongated morphology with greater cell motility in MEFs (Goulimari et al. Citation2005). In order to determine if the elongated, migratory phenotype was also present in the established RKO colon cancer cell line, containing an inducible KLF4 construct, the KLF4-deleted RKO cells were treated with PA or with EtOH as a control for three days prior to visualization. The PA-treated cells expressing KLF4 presented a greater number of protrusions compared to the EtOH-treated cells, which exhibited a more typical compact epithelial shape (). Quantification of the differences revealed that they were statistically significant with a p value of .0036 (). Previous work by Takahashi et al. (Citation2014) has demonstrated that the non-polar morphology in RKO cells was correlated with increased migration. Given the role of actin polymerization in cell morphology and migration, the MEFs were stained with phalloidin to visualize the actin cytoskeleton (). We found no significant difference in the percentage of cells expressing lamellipodia and filopodia between wild-type and Klf4-null MEFs, with p values equal to .7598 and .7481, respectively (). However, there was a significant difference in the presence of stress fibers with a p value equal to .0071. These findings led us to hypothesize that the particular increase in stress fibers in Klf4-null cells may result in more motility-enabled cell populations (Goulimari et al. Citation2005) ().
Figure 1. Klf4 deficient cells exhibit an altered morphology associated with migration when compared to cells expressing Klf4. (A) Representative wild type and Klf4-null MEFs morphology. Early passage cells were imaged using brightfield at 10× magnification. (B) Quantification of migratory morphology in A with n = 9 and p ≤ .0001. (C) Repeat of procedure in A, but using ethanol and PA-treated RKO cells. (D) Quantification of migratory morphology in C with n = 3 and p = .0036. (E) Representative wild type and Klf4-null MEFs stained with phalloidin 555 nm at 40× magnification. (F) Quantification of phalloidin-stained actin structures for wild type and Klf4-null MEFs, n = 3 and p = .0071 (stress fibers), p = .7481 (filopodia), and p = .7598 (lamellipodia). Arrows indicate a migratory phenotype. **Indicates p ≤ .01, *** indicates p ≤ .001. Error bars indicate standard deviation of replicates.
Klf4 null cells exhibit increased rate of migration
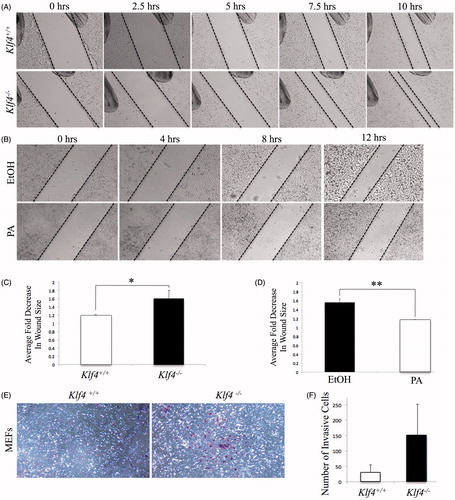
In order to confirm the role of Klf4 in migration, scratch assays were performed with both MEFs and RKO cells. Wild-type and Klf4-null MEFs were plated to confluence, cultured in reduced serum condition to synchronize cell cycles and limit cellular proliferation, and then scratched. The change in wound area caused by the cells migrating into the empty space, relative to a reference mark, was documented via brightfield microscopy at specific time points after generation of the wound (). Klf4−/− MEFs demonstrated increased wound closure compared to wild-type over the time period with a p value equal to .041 (). After treatment with PA and EtOH under reduced serum conditions, the RKO cells were scratched, following the identical procedure as the MEFs (). Comparing 0 hours to 12 hours post scratch indicated that the RKO cells treated with EtOH were more migratory compared to those treated with PA. The quantification of the difference in migration was statistically significant with a p value of .0028 (). Our findings are consistent with previously published data involving colon cancer cell model that demonstrated overexpression of Klf4 inhibits migration (Dang et al. Citation2003). Here, we demonstrate in a non-transformed, MEF cell line that Klf4 expression inhibits migration ().
Figure 2. Klf4 deficient cells present with increased migration as well as invasive trend compared to cells expressing Klf4. (A) Wild type and Klf4-null MEF monolayer scraped with a pipette in 1% serum starvation and imaged at 4× magnification. (B) Ethanol and PA treated RKO cell monolayer in 1% serum starvation imaged at 4× magnification. (C) Quantification of averaged fold decrease in area of A between t = 0 and t = 7.5, n = 5 with a p value of .041. (D) Quantification of averaged fold decrease in area of B between t = 0 and t = 12, n = 3 with a p value of .0028. * indicates p ≤ .05, ** indicates p ≤ .01. (E) Matrigel invasion assay after 72-hour incubation period, after which invasive cells were stained and documented via brightfield microscopy at 10× magnification. (F) Quantification of number of cells present in each respective well; n = 2 for MEFs and p =.196. Error bars indicate standard deviation of replicates.
Klf4 deficient cells show a greater invasion trend compared to cells expressing Klf4
To determine the invasive capacity of cells null for Klf4, an invasion assay utilizing matrigel was performed. MEFs were seeded in the matrigel invasion assay wells and incubated for 72 hours. Documentation of Crystal Violet-stained membranes through brightfield microscopy revealed that Klf4 null cells have higher invasive potential (). Quantification of all the invasive cells in the wells showed an average of 152 Klf4−/− MEFs were capable of passing through the synthetic membrane into 10% FBS supplemented media. In contrast, only an average of 32 Klf4+/+ displayed an invasive phenotype (), showing almost a fivefold increase with loss of Klf4. The trend, of MEFs lacking Klf4 showing increased invasive capability, is consistent with previous findings demonstrated in RKO cells (Dang et al. Citation2003). However, our invasion assays carried high variability, which may have been due to the absence of stronger chemoattractant across the basal polycarbonate membrane. In vivo, metastasis is driven by chemoattraction, to which cells with invasive phenotypes can respond, an aspect our assay is lacking (Zhou et al. Citation2014; Orellana et al. Citation2015). The observed data in combination with Dang et al. (Citation2003) suggests the ability of cells lacking Klf4 to more easily cross the matrigel membrane due to the function of KLF4 in inhibiting invasion.
KLF4 overexpression reduces stress fiber formation in Klf4−/− MEFs
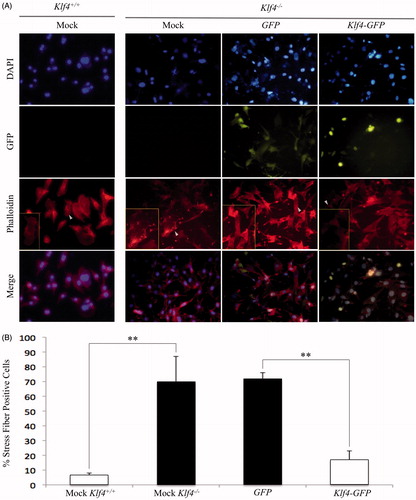
To confirm the percentage of cells expressing migration-associated stress fibers, mock transfected MEFs wild type or null for Klf4 were co-stained with phalloidin and DAPI (). Visualization by fluorescent microscopy of mock transfected Klf4−/− MEFs revealed a statistically significant increased level of stress fibers, 68.2%, compared to mock transfected Klf4+/+ MEFs, 7.1%. The calculated p value was equal to .00731 (). To confirm the function of Klf4 in stress fiber formation, Klf4-deficient MEFs were either transfected with plasmids GFP or GFP conjugated to Klf4 before being fixed and stained with phalloidin and DAPI (). Scoring cells based on the presence or absence of stress fibers indicated that 71.5% of GFP-transfected MEFs expressed the migratory phenotype compared to only 17% of Klf4-GFP transfected MEFs (). The difference in stress fibers between GFP and Klf4-GFP transfected cells was statistically significant with p equal to .0097. The similar rate of stress fibers for Klf4 null MEFs and GFP-transfected MEFs compared to Klf4 wild-type cells and Klf4−/− cells transfected with Klf4-GFP suggests that Klf4 is sufficient to regulate stress fibers ().
Figure 3. Reintroduction of Klf4 reduces stress fiber formation in Klf4-null MEFs. A) Representative images of DAPI and Phalloidin 555 stained images at 20× magnification. Arrows indicate boxed in cells showing representative cells of the population. (B) Quantification of stress fibers in A with n = 3 and p = .0097 for Klf4-GFP and GFP-transfected MEFs and n = 3 and p = .00731 for mock wildtype and mock Klf4-null transfected MEFs. ** indicates p ≤ .01. Error bars indicate standard deviation of replicates.
Lack of Klf4 does not alter RhoA or RhoC gene expression
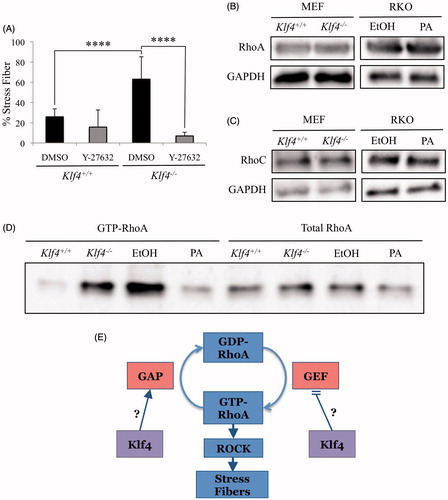
Taken together, our data and previous research demonstrates that Klf4 inhibits motility-associated phenotypes. We therefore wanted further insight into the mechanism by which Klf4 mediates the dynamics of the actin cytoskeleton. Since Rho GTPases are known to coordinate the dynamics of the actin cytoskeleton, our efforts were focused on identifying a potential relationship between KLF4 and Rho GTPases (Ridley Citation2013; Lei et al. Citation2014). We began by testing the expression of RhoA/B/C family proteins with an antibody that targets a shared epitope, finding increased RhoA/B/C expression in Klf4−/− compared to Klf4+/+ (data not shown). Further testing with RhoA and RhoC was selected following a decrease in stress fibers after treatment with ROCK inhibitor (), which is a key effector in signal transduction of the RhoA and RhoC pathways in stress fiber formation and motility (O’Connor and Chen Citation2013). RhoB was not considered for testing due to its role in cell survival and not motility (Wheeler and Ridley Citation2004; Ridley Citation2013). Probing with RhoA () and RhoC () specific antibodies demonstrated no discernible difference in relative protein expression. Our findings are consistent with prior microarray data as no differences were observed between Klf4+/+ and Klf4−/− MEFs for RhoC at the mRNA level (Hagos et al. Citation2011). However, this microarray data identifies upregulated expression of RhoA mRNA, a difference that does not translate to protein expression as shown in our western analysis. The lack of a translational regulation by KLF4 is also supported with previously published ChIP-seq data for KLF4 demonstrating no significant binding score between KLF4 and regulatory regions controlling RhoA or RhoC expression (Chen et al. Citation2008) ().
Figure 4. Klf4 deficient cells have elevated GTP-RhoA. (A) Quantification of phalloidin acti-stain 555 and DAPI co-stained MEFs, either wildtype or Klf4-null, treated with Y-27632 for 24 hours at 10μM; n = 3. **** indicates p < .0001. (B) Representative images of western blots. RhoA primary (21 kDa) antibody, GAPDH (37 kDa) antibody. (C) Representative western blots hybridized with RhoC (19 kDa) antibody, GAPDH (37 kDa) antibody. (D) Representative RhoA activity assay and western blotting of lysate from the RhoA activity assay prior to GTP-RhoA isolation with n = 3. (E) Proposed mechanism of Klf4 in stress fiber formation. Error bars indicate standard deviation of replicates.
Klf4-deficient cells exhibit increased RhoA activity as demonstrated by upregulated GTP-RhoA
RhoA is a GTPase that has been shown to induce migration via stress fiber formation when activated (Chen et al. Citation2014). RhoA activity is regulated by its binding to GTP or GDP, which is in turn regulated by GAP and GEF (Boguski and McCormick Citation1993; Van Aelst and D’Souza-Schorey Citation1997). The relationship raises the possibility that RhoA regulation may happen posttranslationally rather than through the gene expression. In order to examine if the presence or absence of Klf4 correlates with GTP-bound RhoA, we assayed the binding of RhoA to the Rho-binding domain of Rhotekin, a Rho effector protein with a high affinity for the GTP-bound form of RhoA (Ren et al. Citation1999). MEFs and RKO cells were starved at 1% FBS to maintain a controlled state prior to RhoA pulldown and subsequent western blotting. We found that both Klf4−/− MEFs and RKO cells lacking KLF4 expression exhibited elevated levels of GTP-RhoA compared to Klf4+/+ MEFs and KLF4-induced RKO cells (). Quantification of these results reveals a fourfold elevation in RhoA activity in both RKO and MEF cells lacking Klf4 expression (data not shown). Our findings are consistent with previous work where GTP-RhoA is increased in MEFs that are more migratory (Goulimari et al. Citation2005). The elevated levels in the absence of Klf4 expression indicate a possible mechanism in which Klf4 regulates either GEF or GAP to alter RhoA activity. To increase GTP-bound RhoA, either GEF may be upregulated, or GAP downregulated. Our data suggests that in wild-type cells, KLF4 either inhibits GEF or activates GAP to decrease the level of GTP-RhoA, such that a decrease in the rate of stress fiber formation, associated with migration, would be observed (). Compatible with our findings, the ChIP-seq data present numerous GEFs and GAPs to whose regulatory regions KLF4 significantly binds (Chen et al. Citation2008).
Future work is required to determine the specific GEF or GAP that is transcriptionally regulated by Klf4. Luciferase assays could be used to determine direct KLF4 binding to GEF or GAP regulatory gene sequences. Expanding our understanding of the molecular mechanism may prove valuable in better describing the metastatic process and identifying specific druggable targets further downstream. Designing a drug to prevent migration and invasion of Klf4 negative cancers, by targeting selected components of the pathway, could enhance the effectiveness of current cancer treatments.
Acknowledgements
We acknowledge the lab of Dr Vincent W. Yang (Stony Brook Medical School, NY) for providing KLF4-GFP and GFP constructs. The authors would also like to thank Dr Kenneth Belanger for critically reviewing the manuscript.
Disclosure statement
All authors declare that they have no competing interests at this point of time.
Additional information
Funding
References
- Boguski MS, McCormick F. 1993. Proteins regulating ras and its relatives. Nature. 366:643–654.
- Chen X, Johns DC, Geiman DE, Marban E, Dang DT, Hamlin G, Sun R, Yang VW. 2001. Krüppel-like Factor 4 (Gut-enriched Krüppel-like Factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 276:30423–30428.
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 133:1106–1117.
- Chen X, Corbin JM, Tipton GJ, Yang LV, Asch AS, Ruiz-Echevarria MJ. 2014. The TMEFF2 tumor suppressor modulates integrin expression, RhoA activation and migration of prostate cancer cells. Biochim Biophys Acta Res. 1843:1216–1224.
- Dang DT, Mahatan CS, Dang LH, Agboola IA, Yang VW. 2001. Expression of the gut-enriched Krüppel-like factor (Krüppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on CDX2. Oncogene. 20:4884–4890.
- Dang DT, Chen X, Feng J, Torbenson M, Dang LH, Yang VW. 2003. Overexpression of Krüppel-like factor 4 in the human colon cancer cell line RKO leads to reduced tumorigenicity. Oncogene. 22:3424–3430.
- El-Karim EA, Hagos EG, Ghaleb AM, Yu B, Yang VW. 2013. Krüppel-like factor 4 regulates genetic stability in mouse embryonic fibroblasts. Mol Cancer. 12:89.
- Garrett-Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B. 1996. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem. 271:31384–31390.
- Goulimari P, Kitzing TM, Knieling H, Brandt DT, Offermanns S, Grosse R. 2005. Galpha12/13 is essential for directed cell migration and localized Rho-Dia1 function. J Biol Chem. 280:42242–42251.
- Hagos EG, Ghaleb AM, Dalton WB, Bialkowska AB, Yang VW. 2009. Mouse embryonic fibroblasts null for the Krüppel-like factor 4 gene are genetically unstable. Oncogene. 28:1197–1205.
- Hagos EG, Ghaleb AM, Kumar A, Neish AS, Yang VW. 2011. Expression profiling and pathway analysis of Krüppel-like factor 4 in mouse embryonic fibroblasts. Am J Cancer Res. 1:85–97.
- Huang CC, Liu Z, Li X, Bailey SK, Nail CD, Foster KW, Frost AR, Ruppert JM, Lobo-Ruppert SM. 2005. KLF4 and PCNA identify stages of tumor initiation in a conditional model of cutaneous squamous epithelial neoplasia. Cancer Bio Ther. 4:1401–1408.
- Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. 2002. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 129:2619–2628.
- Lehtimaki J, Hakala M, Lappalainen P. 2017. Actin filament structures in migrating cells. Handb Exp Pharmacol. 235:123–152.
- Lei R, Tang J, Zhuang X, Deng R, Li G, Yu J, Liang Y, Xiao J, Wang HY, Yang Q, Hu G. 2014. Suppression of MIM by microRNA 182 activates RhoA and promotes breast cancer metastasis. Oncogene. 33:1287–1296.
- Li J, Zheng H, Yu F, Yu T, Liu C, Huang S, Wang TC, Ai W. 2012. Deficiency of the Kruppel-like factor KLF4 correlates with increased cell proliferation and enhanced skin tumorigenesis. Carcinogenesis. 33:1239–1246.
- Liu C, La Rosa S, Hagos EG. 2014. Oxidative DNA damage causes premature senescence in mouse embryonic fibroblasts deficient for Krüppel-like factor 4. Mol Carcinog. 54:889–899.
- Liu C, Deroo EP, Stecyk C, Wolsey M, Szuchnicki M, Hagos EG. 2015. Impaired autophagy in mouse embryonic fibroblasts null for Krüppel-like Factor 4 promotes DNA damage and increases apoptosis upon serum starvation. Mol Cancer 14:101.
- Liu L, Wang Y, Yu Q. 2014. The PI3K/Akt signaling pathway exerts effects on the implantation of mouse embryos by regulating the expression of RhoA. Int J Mol Med. 33:1089–1096.
- Lv H, Zhang Z, Wang Y, Li C, Gong W, Wang X. 2016. MicroRNA-92a promotes colorectal cancer cell growth and migration by inhibiting KLF4. Oncol Res. 23:283–290.
- O’Connor K, Chen M. 2013. Dynamic functions of RhoA in tumor cell migration and invasion. Small GTPases. 4:141–147.
- Ohnishi S, Ohnami S, Laub F, Aoki K, Suzuki K, Kanai Y, Haga K, Asaka M, Ramirez F, Yoshida T. 2003. Downregulation and growth inhibitory effect of epithelial-type Krüppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem Biophys Res Commun. 308:251–256.
- Orellana R, Kato S, Erices R, Bravo ML, Gonzalez P, Oliva B, Valdivia A, Ibañez A, Brañes J, Barriga MI, et al. 2015. Platelets enhance tissue factor protein and metastasis initiating cell markers, and act as chemoattractants increasing the migration of ovarian cancer cells. BMC Cancer. 15:290.
- Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M, Chhieng DC, Grizzle WE, Engler JA, Krontiras H, et al. 2004. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res. 10:2709–2719.
- Pollard TD, Borisy GG. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465.
- Raftopoulou M, Hall A. 2004. Cell migration: Rho GTPases lead the way. Dev Bio. 265:23–32.
- Ren X, Kiosses WB, Schwarz MA. 1999. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18:578–585.
- Ridley AJ, Hall A. 1992. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 70:389–399.
- Ridley AJ. 2013. RhoA, RhoB and RhoC have different roles in cancer cell migration. J Microsc. 251:242–249.
- Rowland BD, Peeper DS. 2006. KLF4, p21 and context-dependant opposing forces in cancer. Nat Rev Cancer. 6:11–23.
- Shields JM, Christy RJ, Yang VW. 1996. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J Biol Chem. 271:20009–20017.
- Takahashi R, Nagayama S, Furu M, Kajita Y, Jin Y, Kato T, Imoto S, Sakai Y, Junya T. 2014. AFAP1L1, a novel associating partner with vinculin, modulates cellular morphology and motility, and promotes the progression of colorectal cancers. Cancer Med. 3:759–774.
- Thievessen I, Fakhri N, Steinwachs J, Kraus V, McIsaac RS, Gao L, Chen BC, Baird MA, Davidson MW, Betzig E, et al. 2015. Vinculin is required for cell polarization, migration, and extracellular matrix remodeling in 3D collagen. FASEB J. 29:4555–4567.
- Todaro GJ, Green H. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 17:299–313.
- Van Aelst L, D’Souza-Schorey C. 1997. Rho GTPases and signaling networks. Genes Dev. 11:2295–2322.
- Wei D, Gong W, Kanal M, Schlunk C, Wang L, Yao JC, Wu TT, Huang S, Xie K. 2005. Drastic down-regulation of Krüppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 65:2746–2754.
- Wheeler A, Ridley A. 2004. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp Cell Res. 301:43–49.
- Yamazaki D, Kurisu S, Takenawa T. 2005. Regulation of cancer cell motility through actin reorganization. Cancer Sci. 96:379–386.
- Yori J, Seachrist D, Johnson E, Lozada K, Abdul-Karim F, Chodosh L, Schiemann W, Keri R. 2011. Krüppel-like factor 4 inhibits tumorigenic progression and metastasis in a mouse model of breast cancer. Neoplasia. 3:601–610.
- Zammarchi F, Morelli M, Menicagli M, Di Cristofano C, Zavaglia K, Paolucci A, Campani D, Aretini P, Boggi U, Mosca F, Cavazzana A, et al. 2010. KLF4 is a novel candidate tumor suppressor gene in pancreatic ductal carcinoma. Am J Pathol. 178:361–372.
- Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. 2004. Identification of Krüppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 23:395–402.
- Zhou J, Xiang Y, Yoshimura T, Chen K, Gong W, Huang J, Zhou Y, Yao X, Bian J, Wang JM. 2014. The role of chemoattractant receptors in shaping the tumor microenvironment. BioMed Res Int. 2014:751392.

