 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
We report the identification and characterization of a novel gene, AtHesperin (AtHESP) that codes for a deadenylase in Arabidopsis thaliana. The gene is under circadian clock-gene regulation and has similarity to the mammalian Nocturnin. AtHESP can efficiently degrade poly(A) substrates exhibiting allosteric kinetics. Size exclusion chromatography and native electrophoresis coupled with kinetic analysis support that the native enzyme is oligomeric with at least 3 binding sites. Knockdown and overexpression of AtHESP in plant lines affects the expression and rhythmicity of the clock core oscillator genes TOC1 and CCA1. This study demonstrates an evolutionary conserved poly(A)-degrading activity in plants and suggests deadenylation as a mechanism involved in the regulation of the circadian clock. A role of AtHESP in stress response in plants is also depicted.
Introduction
Circadian rhythms are ubiquitous time-keeping phenomena in eukaryotes with a period of ∼24 h that represent an adaptive strategy to daily environmental changes. Three main components are recognized in circadian systems: the input pathways, the central oscillator and the output pathway. The basic mechanism of the oscillator is well conserved and a general principle of autoregulatory, interconnected feedback loops of “core” gene transcription and translation applies that further regulate the expression of downstream targets.Citation1-3 In Arabidopsis thaliana, 3 genes, LHY, CCA1 and TOC1, regulate the master oscillator via a well-described repressive mechanism at transcriptional level, which has recently expanded with numerous components.Citation4 Further, post-transcriptional and post-translational regulations, and chromatin remodeling are critical for maintaining the circadian rhythmicity in eukaryotic clocks.Citation5-7 Among these regulatory mechanisms, the stability and the turnover rates of mRNAs involved in the circadian system are key issues in transcript oscillations.Citation8 For example, in flies, the daily oscillation of per (a central oscillator gene) depends on its mRNA life time and stability Citation9; in mammals, hnRPN and PTB1 bind the 3′-UTR of Cryptochrome1 and Period2 causing their degradation Citation10,11; in Neurosporacrassa the clock-controlled rrp44 gene, coding for the 3′→5′ exonuclease subunit of exosome, is implicated in the degradation of the circadian regulated frq mRNA Citation12; in plants, the light-regulated transcript levels of the CCA1 “core gene,” and the oscillation of various “output” genes are ascribed to the stability of their mRNAs,Citation13,14 although the steps and the factors that regulate mRNA turnover of circadian genes remain essentially unknown.
The first and rate-limiting step of cytoplasmic mRNA turnover is the shortening of poly(A) tails, catalyzed by Mg2+-dependent, 3′→5′ exoribonucleases, known as deadenylases.Citation15,16 They are classified to the DEDD and the exonuclease-endonuclease-phosphatase (EEP) superfamiliesCitation16; the former are named after conserved catalytic Asp and Glu residues, while the latter have conserved Asp and His residues in their active site.Citation16 The main deadenylase complexes are CCR4-NOT (Carbon Catabolite Repressor 4 – Negative on TATA), PARN [poly(A)-specific ribonuclease] and PAN [poly(A) nuclease] complexes. Of these, CCR4-NOT and PARN have been studied in Arabidopsis, while studies on PAN are still limited.Citation17-19 CCR4 and the CCR4-CAF1 (CCR4-associated factor 1) consist the major eukaryotic deadenylation complex known in eukaryotes up to date. Among these, Nocturnin (NOC) is a circadian deadenylase, first identified in X. laevis, also present in mammals, yeast and flies.Citation20,21
Although A. thaliana may have up to 26 deadenylases,Citation16 deadenylation in plants has not been extensively studied, while a poly(A)-degrading enzyme associated to the clock is still elusive. Herein, we identify a NOC homolog in Arabidopsis that is transcribed at highest levels in the evening. Due to its expression profile we name it A. thaliana HESPERIN (AtHESP) from the Greek word “Eσπερoς; (Hesperos), the Evening Star. AtHESP is a novel transcriptional regulator of circadian rhythms with poly(A)-degrading activity that affects rhythmically expressed mRNAs of the core oscillator in plants while a negative role in the plant response against oxidative stress is also evident.
Materials and methods
Identification of AtHESP
For mining the A. thaliana genome toward the identification of deadenylase genes, the TBLASTN algorithm of TAIR database (http://www.arabidopsis.org/Blast/index.jsp) was used against the TAIR10 transcripts dataset. The presence of cis-elements present in the promoter regions of genes of interest was investigated using the PLACE plant cis-elements database (http://www.dna.affrc.go.jp/PLACE/index.html).Citation22 A PCR amplicon comprising the full-length cDNA clone of AtHesperin was produced, using cDNA from 7 days old plants as template and a pair of specific primers HESP-F and HESP-R (). The alignment and phylogenetic analysis of the deadenylase protein sequences were performed with MEGA 5.05 software package.Citation23 The multiple alignment parameters were adjusted with gap cost 10 and gap extension 1. The phylogenetic trees were constructed using the neighbor-joining algorithm with bootstrap analysis of 1000 replicates.
Table 1. Primers used in experimental procedures.
Cloning, expression and purification of recombinant AtHESP
For AtHESP overexpression, the complete cDNA sequence was subcloned into pET15b vector and used to transform E.coli cells. Expression was induced at A600 0.6 with 1 mM IPTG and culturing for 4 more hours. The AtHESP polypeptide was purified using Ni-NTA Agarose affinity chromatography matrix (QIAGEN) equipped on an ÄKTApurifier (GE Healthcare) following the manufacturer's instructions.
Size exclusion chromatoghraphy (SEC) analysis
SEC analysis was carried out using a Tricorn Superose 12 10/300 GL column (GE Healthcare) adapted on an ÄKTApurifier (GE Healthcare). The column was pre-equilibrated for 2 column volumes using Buffer A (0.1 M Sodium Phosphate Buffer, pH 6.0, 0.5 M NaCl) with a flow rate of 0.4 ml/min on average. About 500 μl of sample was loaded on the injection valve. Elution was performed using the same Buffer. The absorbance was monitored at A280. The SEC profile was analyzed using the software Unicorn™ (Amersham). Protein concentration was 16 mg/ml. Glucogen phosphorylase from rabbit muscle (97 kDa, 3 mg/ml) and lysozyme (14.3 kDa, 2 mg/ml) were used as size markers.
Assays for poly(A) degradation
The enzymatic activity of recombinant AtHESP was determined by a colorimetric assay as described with some modifications.Citation24-26 Briefly, the reactions were performed with 0.1-0.15 nM recombinant AtHESP in 1 ml reactions. The absorbance of poly(A), poly(U), poly(C) or poly(G) (all from Sigma-Aldrich) was measured at appropriate wavelengths [poly(A), 662 nm; poly(U), 663 nm; poly(G), 665 nm; poly(C), 667 nm]. Deadenylation reactions with in vitro transcribed L3A30 and L3A30X49 RNAs were performed as described elsewhere Citation27,28; reacted RNA was purified, analyzed in 3% agarose gels and developed with GelRed (Biotium) staining.Citation29-31 Polyacrylamide gel electrophoresis under native conditions for acidic proteins (pI < 7) was performed as described previously.Citation32
Kinetic analysis and calculation of binding sites
To calculate the number of the binding sites and the kinetic parameters of AtHESP for poly(A) as substrate several kinetic approaches were used.Citation33 First, Lineweaver-Burk formalism applied for allosteric enzymes that show sigmoidal response in velocity versus substrate plots. Upon application of the formalism in such enzymes the 1/v vs. 1/[S] plot is curved, while a linear plot may be obtained for 1/v versus 1/[S]n, where n is the power that yields a straight line; in this case the procedure yields an apparent n value (napp) and equals to n. Second, the analysis of the data in terms of Hill equation allows the calculation of the Hill coefficient which is used to evaluate the degree of allosteric cooperativity according to the following equation(1)
(1) where nH is the slope and KM′ is the dissociation constant. nH> 1 suggests strong positive kinetic cooperativity and then nH tends to the number of substrate binding sites. KM′ may be calculated using the equation
(2)
(2) where, [S]0.5 is the substrate concentration at 0.5 Vmax.
Additionally, the number of the binding sites can be calculated by the shape of the v - [S] curve and 2 fractions of Vmax, usually 0.9Vmax and 0.1Vmax, according to the equation(3)
(3) where, napp represents the apparent number of binding sites, [S]0.9 and [S]0.1 the substrate concentrations at 0.9Vmax and 0.1Vmax, respectively. The next higher integer above napp represents the minimum number of actual sites (designated as n). However, as the plot is non-linear over the range of velocities between 0.1 to 0.9Vmax, n is determined as the slope of the Hill plot at v = 0.5 Vmax.
Plant material and growth conditions
Arabidopsis thaliana (ecotype Columbia-0) seeds were surface-sterilized and placed at 4°C in the dark for 24 hours in Petri dishes containing Murashige Skoog (MS) medium and 1% agar. They were then transferred at 22°C under a photoperiod of 16/8 h or 12/12 h light/dark cycle, at 150μmol photons m−2s−1. For the evaluation of the photosynthetic parameters, plants were cultivated on a sterilized soil and peat mixture in controlled-environment growth chamber (EF7, Conviron, Montreal, Canada) under long day conditions 16/8 h, with 50 ± 5/60 ± 5% humidity, temperature 22 ± 1/18 ± 1°C and light intensity of 120 ± 20 μmol photons m−2 s−1. All plants were trained for a period of 7 days under stable conditions of temperature and photoperiod. Plant grown under 16/8 h light/dark cycles were sampled every 4 hours from the 8th day on, while in the morning of the 9th day they were transferred in constant light for 2 more days. Plant shoots were collected at certain time points and used for total RNA extraction. Seeds of Arabidopsis SALK insertion lines SALK_037900, SALK_105290 and SALK_127310, were obtained from the European Arabidopsis Stock Center (NASC) and were designated as hesp1-1, hesp1-2, hesp1-3, respectively. Plants genotypes (homozygous or heterozygous for T-DNA insertion) were determined by PCR amplification with gene specific forward and reverse primers (37900-L and 37900-R; 105290-L and 105290- R; 127310-L and 127310-R) and T-DNA primer (LBb1.3) (). F2 seeds from homozygous plant lines were used for circadian expression experiments. Seeds were pregerminated in MS plates and shoots of 8 day-old seedlings were collected at 5 specified time points and used for RNA extraction and qRT-PCR experiments.
Production of plant lines that overexpress AtHESP
The full-length clone of AtHESP was ligated into pCambia35S-eGFPn (pCambia 2201 backbone), giving rise to the pCambia-HESP construct. Agrobacterium tumefaciens AGL1 cells were used for plant transformation by the floral dip method.Citation20 Independent transgenic lines from F2 were obtained on selective MS medium containing kanamycin and transferred into pots with soil in a growth chamber as described above. Genomic DNA was extracted from 16 individual kanamycin resistant 20 days old plants and used in PCR with 35S-F and 35S-R primers () for the identification of positive transgenic lines. Leaves from transgenic plant lines 30 days old were collected at 2 time points (in the morning and just before transition to the dark) and used for RNA extraction and Q-PCR experiments.
Chlorophyll fluorescence measurements
Chlorophyll fluorescence was measured at room temperature in the leaves of 4-week-old A. thaliana Col-0 and hesp1-1 knock down mutants, using an imaging-PAM fluorometer (Walz, Effeltrich, Germany), as described before.Citation34 All leaves were dark adapted for 15 min before measurement. Five areas of interest (AOI) were selected, one in the center of the leaf, 2 in the outer zone of the tip and 2 in the base of the leaf. First Fo (minimum chla fluorescence in the dark) and Fm (maximum chla fluorescence in the dark) values were measured with dark-adapted samples. Fm was obtained with a saturating pulseCitation34 of white light (2400 μmol photons m−2 s−1, 800 ms) followed by application of actinic light to assess steady-state photosynthesis (Fs). The actinic light of 120 μmol photons m−2 sec−1 was selected to match that of the growth light of A. thaliana plants, and low enough to avoid photo inhibition. The illumination time was 3 min with measurement of Fo′(minimum chla fluorescence in the light) and Fm′ (maximum chla fluorescence in the light) from which, automatically values of other chl fluorescence parameters were calculated by the Imaging Win software. The value of Fo′was estimated using the approximation of Oxborough and Baker.Citation35 The calculated parameters include the effective quantum yield of photochemical energy conversion in PSII (ΦPSII), that estimates the efficiency at which light absorbed by PSII is used for photochemistry, the yield of regulated non-photochemical energy loss in PSII (ΦNPQ), that is the quantum yield for dissipation by down regulation in PSII and ΦNO, the quantum yield of non-regulated energy loss in PSII.Citation36 Photochemical quenching (qp), a measure of the fraction of open PSII reaction centers, that is the redox state of quinone A (QA), primary acceptor of PSII, and the non-photochemical quenching (NPQ), that estimates heat dissipation of excitation energy were also calculated. The relative PSII electron transport rate (ETR) was calculated according to Schreiber et al.Citation37 Excitation pressure, measured as 1-qp, is an estimate of the proportion of closed PSII reaction centers.Citation38
Real time PCR. Shoots from plants, collected at time points as described above, were grounded in liquid nitrogen
Total RNA was isolated from 15 plants for each one of 3 biological repeats in every time point using RNeasy extraction Kit (QIAGEN). To eliminate genomic DNA contamination, RNA samples were treated with DNaseI (Invitrogen) according to manufacturer's instructions. First strand cDNA was reverse transcribed with SuperScript II™ Reverse Transcriptase (Invitrogen) according to instructions provided by the manufacturer. The resulting first-strand cDNA was normalized for the expression of housekeeping gene of Ubiquitin (UBQ). Gene specific primers were designed for the HESP, UBQ, CCAI TOCI and LHY genes with Beacon designer v7.01 software (). Quantitative RT-PCR reactions were performed on MX 3005P system (Stratagene, CA, USA) using KAPASYBR® FAST qPCR (Kapa Biosystems) and gene-specific primers following the manufactures instructions. PCR cycling started with the initial polymerase activation at 95°C for 5 min, followed by 45 cycles of 95°C for 15 sec, 58°C for 20 sec and 72°C for 10 sec. The primer specificity and the formation of primer-dimers were monitored by dissociation curve analysis and agarose gel electrophoresis on a 3 % (w/v) gel. In all samples a single amplicon has been detected. Relative transcript levels in different samples for the gene of interest were calculated as a ratio to the UBQ gene transcripts. Data were analyzed according to PfafflCitation39 and the reactions efficiency have been estimated with LinReg PCR.Citation40 For all samples from different time points, 3 biological replicates of PCR reaction were performed for each gene.
Statistical analysis
All experiments were conducted at least twice and analyzed by analysis of variance Citation17 followed by Duncan multiple comparison tests (a < 0.05). Chlorophyll fluorescence measurements represent averaged values (n = 6) from 2 independent experiments with 3 leaf samples (each with 5AOI) from 3 different plants, per treatment per experiment. Standard errors were calculated for all mean values and t-tests were performed for pair wise comparisons of means at different time points (P ≤ 0.05).
Results
AtHESP gene codes for a deadenylase in A. thaliana
In order to identify circadian deadenylase genes in plants, Xenopus laevis Nocturnin (NOC) amino acid sequence Citation21 was used as query sequence for mining the A. thaliana genome. At least 8 different genes with homology to NOC were identified. As NOC exhibits circardian expression, the promoter regions (up to 2 kb upstream) of the putative orthologswere searched for the presence of cis-elements previously reported to be implicated in circadian regulation of gene expression.Citation41,42 Among these, only in the promoter of At1G31500 we identified: a) 2 conserved Evening Element (EE) regulatory sequences (AAATATCT) at positions −707 and −873, previously characterized in A. thaliana,Citation43,44 b) B type E-boxes, CACGTG, also called G-boxes in plants,Citation45 c) 3 palindromic hexanucleotide E-box elements (CANNTG) at positions −328 (CAGGTG), −399 (CAATTG) and −482 (CAAATG), respectively, similar to those reported in other NOC promoters from Mammalia, vertebrates, chicken and insects,Citation46,47 and d) 4 CCA1 binding sites (AATCT) at positions −149, −277, −343 and −424 binding elements, characterized as CCA1/LHY binding sites on TOC1 promoter Citation43,48 (). Thus, AT1G31500 gene, named AtHesperin (AtHESP), was identified as a putative NOC homolog in A. thaliana.
Figure 1. (A). Analysis of AtHESP (At1G31500) promoter. EEs, CCA1 Binding Sites and E-boxes important for circadian regulation are marked. (B). Schematic representation of AtHESP (417 amino acids) showing the conserved nuclease domain (in blue) related to the Mg2+-dependent EEP superfamily, in comparison with the mouse NOC and human orthologHsCNOT6L. The latter possesses N-terminal leucine-rich repeats (LRRs) (shown in gray). The amino acid lengths are given for each polypeptide. The positions of the conserved catalytic amino acids are also indicated. (C). Partial AtHESP amino acid sequence alignment with characterized NOCs, focusing on conserved amino acid residues. HsNOC, MmNOC, RnNOC and XlNOC, orthologs of the EEP deadenylase family (HsCNOT6, HsCNOT6L and ScCCR4), and plant orthologs RcNGL1 and PtEEP, are included. The black line indicates the conserved PDILCLQEV domain for Mg2+/Mn2+ binding, which includes the catalytic residues E193 of MmNOC or E556 of ScCCR4 (both indicated by filled polygon). The filled square, the filled ellipse and the open star indicate catalytic residues D713, D780 and H818 of ScCCR4, respectively. Colors of residues are according to amino acid polarity. (D). Phylogenetic analysis of NOC, CCR4 and EEP protein sequences. The percentage of replicate trees in which the associated taxa clustered together is shown next to the branches. Scale bar, 0.1 substitutions per site. Hs, Homo sapiens; Ms, Mus musculus; Pt, Populus trichocarpa; Rc, Ricinus communis; Rn, Rattus norvegicus; Sc, Saccharomyces cerevisiae; Xl, Xenopus laevis.
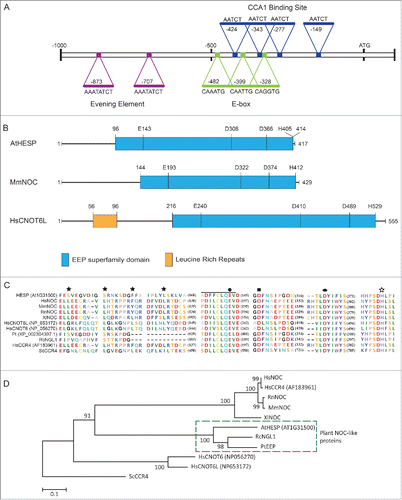
The deduced amino acid sequence of AtHESP results to a theoretical molecular mass of 47.1 kDa and isoelectric point of 6.80. AtHESP, like NOC, possess high similarity to the catalytic C-terminus of ScCCR4 and the catalytically important residues E556, D713 and D780 and H818 (residues E143, D308, D366 and H405) Citation49 of ScCCR4 as well as the PDILCLQEV domain involved in Mg+2 or Mn+2 binding (positions 136-145) Citation50 are conserved in all deadenylases aligned, including AtHESP (). The sequence was compared with the ones of biochemically characterized NOCs from human (HsNOC; GenBank accession number NP036250),Citation50 mouse (MmNOC; NP033964),Citation51 Xenopus laevis (XlNOC; NP001079281) Citation21 and Rattus norvegicus (RnNOC; NP612535),Citation51 as well as with CCR4 members of CCR4-NOT deadenylation complex, conserved among the organisms, namely the human HsCNOT6 (CCR4a; NP056270), HsCNOT6L (CCR4b; AF183961) and yeast Saccharomyces cerevisiae ScCCR4 (AAB24455).Citation52-54 Plant orthologues with a high similarity to AtHESP were also identified in the databases from Ricinus communis (RcNGL1; XP002519903) and Populus trichocarpa (PtEEP; XP002304397) and included in the analysis (). The analysis showed that AtHESP is not very closely related to characterized NOC proteins sharing 23%, 22.6%, 22.8% and 23% similarity to XlNOC, MmNOC, HsNOC and RnNOC, respectively.
Our in silico analysis suggests that AtHESP codes for a putative deadenylase sharing similarities to previously characterized EEP deadenylases, including the circadian regulated NOCs.
AtHESP is a Mg+2-dependent poly(A)-specific 3′ nuclease. Based on our in silico analysis, we evaluated the ability of purified recombinant AtHESP (Fig. S1) to accept poly(A) as substrate, using a standard colorimetric RNase assay, previously used for kinetic studies on PARN deadenylase.Citation24,55 The plots of the reaction rates vs. poly(A) concentration at several pH values and temperatures produced saturation curves, suggesting that AtHESP can efficiently degrade poly(A) (). Optimal conditions for degradation were at 25°C, pH 6.5 (compare the reaction rate values shown at left and right panels in ).
Figure 2. AtHESP is a Mg2+-dependent poly(A)-specific 3′ nuclease. (A). AtHESP degrades poly(A). Reaction rates of poly(A) degradation by AtHESP were measured using a colorimetric assay at the indicated substrate concentrations at 25°C (left panel) and 30°C (right panel), at 3 pH values (6.5, rhombus; 7.0, square; 7.5, circle). (B). AtHESP activity depends on Mg(II) ions. The rates in the absence (0mM), or excess (5mM) of Mg(II) were compared to those of 1.5 mM considered as the control. Mean values of 3 independent experiments. (C). Specificity for poly(A). The rates for poly(U), poly(C) and poly(G) were compared to those of poly(A). The reactions were performed with 600 mM of each polynucleotide at 25°C, pH 6.5, and 1.5 mM Mg2+. (D). AtHESP is a poly(A)-specific 3′ nuclease. L3A30 (filled arrow) or L3A30X49 (filled double arrow) RNAs were incubated with AtHESP (lanes 3-6) or PARN (lanes 7-10). The reactions were terminated at the indicated time points, denoted by numbers above the lanes (in minutes). The reacted RNA was purified and fractionated on 3% agarose gels stained with GelRed. Numbers on the left and the right indicate the positions of RNA length markers (nt). Filled arrow and double arrow indicate the positions of L3A30 (length 84 nt) and L3A30X49 (length 133 nt) RNA. Open arrowhead indicates the position of the reaction product.
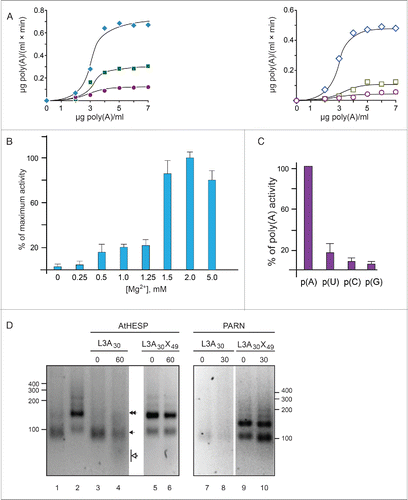
The majority of poly(A)-degrading enzymes depend on Mg(II) for efficient activity.Citation16 Accordingly, assays with [Mg+2] varying from 0.25 mM to 5 mM were performed. The concentration of Mg(II) that showed the highest poly(A)-degrading rate was considered as control to normalize the activity data. shows that AtHESP activity depends on magnesium ions and the optimal Mg(II) concentration for poly(A) degradation is at 2 mM.
Next, we investigated the specificity of AtHESP for RNA substrates, testing the use of poly(U), poly(C), or poly(G), and compared the rates to those of poly(A). Poly(U) was degraded to some extent [∼20% of poly(A)], while poly(C) and poly(G) were almost unaffected (). Thus, AtHESP shows strong specificity for polyadenosine stretches.
To provide further evidence that AtHESP behaves as a poly(A)-specific nuclease, we incubated AtHESP with 2 different RNA substrates previously used for the characterization of PARN deadenylase; L3A30, that bears a stretch of 30 adenosines (A30) at the 3′end of the substrate, and L3A30X49, where the poly(A) stretch is internal followed by an RNA sequence of 49 nucleotides (X49, where X denotes any nucleotide) encoded by the vector.Citation56 Incubation of L3A30 RNA with AtHESP revealed reaction products that correspond to shortened RNAs, while such products were absent in reactions with L3A30X49 RNA (). Since the L3A30X49 RNA pattern was almost unaffected, AtHESP is neither 5′-exoribonuclease, nor endoribonuclease and can only degrade RNA substrates that bear a polyadenosine stretch at their 3′ ends (). To obtain additional evidence for the poly(A)-degrading activity of AtHESP, the electrophoretic profile of the 2 RNAs by AtHESP was compared to that of PARN. shows that the reaction pattern of L3A30 and L3A30X49 RNAs upon incubation with PARN is comparable to the one observed for AtHESP (). We also incubated L3A30 RNA with either AtHESP or PARN for several time intervals, and the reaction products were analyzed on RNA polyacrylamide – urea gels and visualized with silver staining. Both AtHESP and PARN produced analogous patterns of substrate degradation (Supplementary Figure S2A). Finally, we incubated digoxygenin-labeled L3A30 RNA in the presence of AtHESP as described above. The reaction products were fractionated on polyacrylamide gels and faster migrating products were observed upon incubation with AtHESP (Supplementary Figure S2B).
Collectively, the previous results show that AtHESP can efficiently degrade poly(A) stretches in vitro in a Mg(II)-dependent manner, representing a bona fide 3′-end poly(A) exoribonuclease.
Kinetic behavior and evidence for oligomerization
The sigmoid curves of the v - [S] plots obtained in suggest positive cooperativity and are reminiscent of allosteric behavior with multiple catalytic sites. Thus, we analyzed our kinetic data to calculate the potential number of binding sites n, based on several kinetic approaches.Citation33 First we applied Lineweaver-Burk formalism for sigmoidal response curves. In this case, a typical double reciprocal 1/v - 1/[S] plot (where [S] is raised to power 1) produces a curved line, while it is possible to obtain a straight line when [S] is raised to various powers; the n value of the potential binding sites would be the power that yields a straight line in a 1/v vs. 1/[S]n plot. shows that the minimal power that produced a linear 1/v - 1/[S]n plot was n = 3. It should be noted that this approach indicates the minimal potential binding sites. From this formalism, we also calculated the basic kinetic parameters of the enzyme; Vmax equals to 0.086 μg poly(A)/(ml × min) and KM′ to 0.06 μg poly(A)/ml (). Second, the slope nH of the log[v/(Vmax- v)] - log[S] plot (Hill plot) equals to 2.2 (). Values of nH > 1 suggest strong positive kinetic cooperativity during the enzyme reaction, while nH< 1 suggests negative cooperative binding. When the cooperativity is strong, nH also tends to the number of substrate binding sites, suggesting that the next higher integer, n = 3, represents the minimum number of actual sites.Citation33 Finally, we calculated the napp value of the [S]0.9/ [S]0.1 ratio from the sigmoid lines as the one produced at optimal conditions (25°C/pH 6.5; ); the analysis yielded napp = 2.7, suggesting that the enzyme consists of 3 potential substrate sites (the next higher integer above napp). Taken together, our kinetic analyses based on 3 methods suggest that AtHESP shows strong cooperativity with at least 3 substrate sites.
Figure 3. AtHESP is oligomeric. (A, B). Double reciprocal (A) and Hill (B) plots from data obtained from experiments as described in yielded slopes equal to 3 and 2.2, respectively. (C). SEC chromatogram. X axis, elution volume (ml); Y axis, A280 absorbance (arbitrary units; mAu). Asterisks indicate eluted proteins at certain molecular mass (*) 150 kDa, (**) 100 kDa. Arrows indicate the positions of mass markers: single arrow, glucogen phosphorylase (rabbit muscle) 97 kDa; double arrowhead, lysozyme 14.3 kDa. (D). Electrophoretic pattern of SEC fractions under denaturing conditions (10% SDS-PAGE). Numbers in red above the lanes correspond to actual numbering of ÄKTA fractions collected in . Numbers above lanes correspond to actual fraction numbering. Numbers on left indicate position of molecular mass markers (kDa) related to AtHESP size. IN: Input; M: molecular mass markers; E. Electrophoretic pattern of SEC fractions under non-denaturing conditions. Fractions were analyzed in a 7% polyacrylamide gel under non-denaturing conditions. Numbers in red above the lanes correspond to actual numbering of ÄKTA fractions collected in . Urease (Ur) and Bovine Serum Albumin (BA) were used as oligomeric structure and molecular mass markers; urease trimer (272 kDa) and hexamer (545 kDa), and BA monomer (66 kDa) and dimer (132 kDa). Arrows on the left of the gel indicate oligomeric structures, namely dimers, trimers and tetramers. Numbers on the right indicate positions of urease and BA oligomers and monomer.
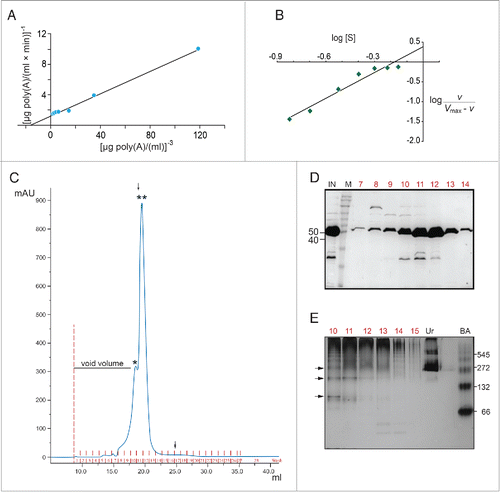
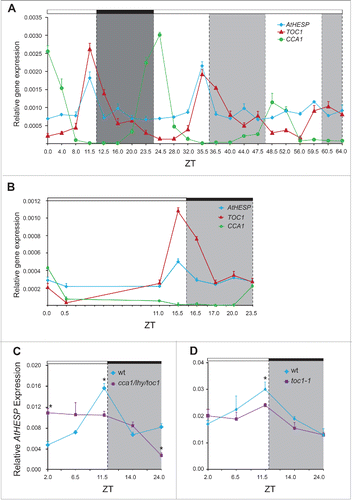
Figure 4. Diurnal and circadian time course of AtHESP, TOC1 and CCA1 expression in A. thaliana plants. A and B. Diurnal time course of expression levels in plants grown under 12 h/12 h light/dark (A) and 16 h/8 h light/dark (Β) photoperiod. Plants in (A) were transferred to constant light at day 7. Transcript levels were recorded every 4 hours. C and D. Diurnal expression of AtHESP in the triple toc1/lhy/cca1 (C) and toc1 mutant line (D) compared to wild-type plants. Plants were grown under 12 h/12 h light/dark photoperiod. White/black bars at the top indicate light-dark conditions, respectively. Light gray frames indicate subjective dark periods. Relative gene expression was measured with respect to internal housekeeping Ubiquitin gene transcripts. Error bars represent standard error of means (n = 3, each consisting of 15 plants). Asterisk (*) indicates statistically different mean values at each time point (t- test P ≤ 0.05). Three independent experimental trials yielded similar results.
To gain additional evidence on the oligomeric stoichiometry of AtHESP, we performed SEC analysis with a Superose 12 10/300 GL column. As the theoretical molecular mass of AtHESP is 47.1, shows 2 major peaks, at approximately 150 and 100 kDa, corresponding to the potential oligomeric states, namely trimeric and dimeric, respectively (). We further analyzed the electrophoretic pattern of the SEC fractions under both denaturing and non-denaturning conditions. The monomeric state of the protein was evident in SDS-PAGE analysis (). The electrophoretic pattern of gel filtration fractions under non-denaturing conditions revealed several bands corresponding to oligomeric forms (), and suggests a stoichiometry of 2 or 3. This is further supported by the patterns observed for urease and bovine serum albumin oligomers (). The pattern in also revealed slower migrating bands that could represent complexes with stoichiometry greater than 3. The kinetic analysis does not exclude this possibility, as linear reciprocal plots were obtained for powers of [S] greater than 3, suggesting n > 3. Taken together the kinetic and SEC analyses suggest that AtHESP is oligomeric, probably with 3 binding sites. Further experimentation is necessary to elucidate the complete structure of the enzyme.
AtHESP is a circadian-regulated gene expressed in the evening
To confirm the anticipated circadian regulation of AtHESP, we examined the AtHesp mRNA levels over diurnal and circadian time courses. In both 12/12 h and 16/8 h light/dark photoperiods, AtHESP exhibits a nearly 3-fold diurnal oscillation in mRNA accumulation with maximum levels just before transition to the dark (). In run-off experiments, where plants growing in 12/12 h light/dark cycle were transferred in constant light for 2 more days, we still observed a rhythmic pattern of AtHESP transcript abundance, suggesting that the transcript itself subjects to circadian regulation (). The two key canonical clock components of the core “central” oscillating loop of Arabidopsis TOC1 (an “evening” gene with maximum of expression before dark) and CCA1 (the “morning” gene with maximum of expression just before dawn), were used as controls. To examine the expression pattern of AtHESP in a disrupted clock-model background, we used the triple cca1/lhy/toc1 mutant plants, where rhythmic responses are expected to be dramatically attenuated.Citation57 In cca1/lhy/toc1 plants, the rhythmicity of AtHESP mRNA expression was significantly affected and its peak appeared at an advanced phase compared to wild type plants (). This anomalous phasing was previously observed in this mutant plant line for other circadian-controlled genes in A. thaliana, namely CAB1, CCR2, GI and PRR5.Citation58,59 On the other hand, the expression of AtHESP in toc1-1 mutant plants remains rhythmic albeit with damped amplitude (). Interestingly, in mutant mice for the core component CLOCK/BMAL1 of the circadian clock, a similar pattern was reported for the expression of Noc.Citation60
Collectively, the previous results show that the expression of AtHESP is regulated by the circadian clock, as has been previously shown for the orthologous genes in other organisms.
AtHESP affects CCA1 and TOC1 expression in Arabidopsis plants
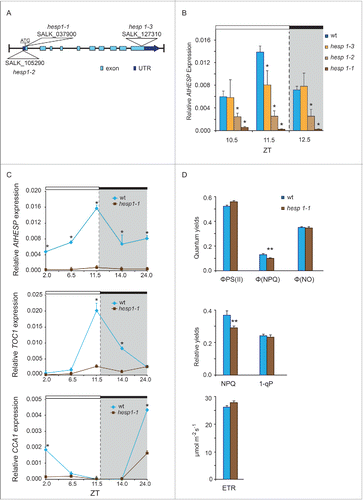
To investigate the function of AtHESP gene in plants, T-DNA homozygous insertion mutants were analyzed for AtHESP expression and compared to Col-0 wild type plants under a 12/12 h light/dark cycle photoperiod. Three homozygous lines hesp1-1 (SALK_037900) hesp1-2 (SALK_105290) and hesp1-3 (SALK_127310) were obtained (). None of the 3 knockdown mutants exhibit any obvious phenotype compared with the wild type plants regarding growth and development under the normal conditions of growth used. In all 3 knock-down mutants, a lack of the rhythmic AtHESP expression is observed (). The diurnal expression of hesp1-1, the most affected knockdown mutant plant line, with highly reduced AtHESP transcription levels, revealed that the circadian oscillation of the gene was abolished (). Interestingly, the expression pattern of CCA1, TOC1 and LHY are also altered in hesp1-1 ( and Supplementary Fig S3). In particular, the expression of TOC1 appears arrhythmic, while the diurnal oscillation and morning expression maximum of CCA1 gene persisted, although in a significantly reduced amplitude. Thus, the regulation of transcript accumulation of clock genes is significantly perturbed in hesp mutants grown in light/dark cycles.
Figure 5. Diurnal gene expression and photosynthetic parameters in A. thaliana knockdown hesp plants. (A). Schematic representation of the insertion positions in the 3 T-DNA homozygous insertion mutants used in the present study. Colored boxes indicate exons and untranslated (UTR) regions. (B). Diurnal time course of AtHESP expression levels in 3 T-DNA homozygous insertion mutants hesp1-1, hesp1-2 and hesp1-3, compared to Col-1 wild type plants.Citation82 (C). Diurnal time course of AtHESP, TOC1 and CCA1 expression in hesp1-1 mutant plants. White/black bars at the top indicate light-dark conditions, respectively. Light gray frames indicate subjective dark periods. Relative gene expression was measured with respect to internal housekeeping Ubiquitin gene transcripts. Error bars represent standard error of means (n = 3, each consisting of 15 plants). (*) Indicates statistically different mean values at each time point relative to wild type (t- test P ≤ 0.05). Three independent experimental trials yielded similar results. D. Assessment of excitation energy flux at PSII in wild-type and hesp1-1 knock down mutant plants estimating the photochemical utilization (ΦPSII), the regulated heat dissipation (ΦNPQ), and non-regulated dissipation (ΦNO) parameters, the non-photochemical fluorescence quenching (NPQ), the excitation pressure (1-qp)representing the fraction of closed PSII reaction centers and the relative PSII electron transport rate (ETR). Plants were grown under 12 h/12 h light/dark photoperiod. Error bars represent standard error of means (n = 6). P values (**<0.01) indicate differences relative to wild type. The experiments were repeated twice.
To further investigate the effect of AtHESP on the expression levels of TOC1 and CCA1, we generated AtHESP overexpressing plant lines (AtHESPox), aiming to examine whether AtHESP acts directly on the TOC1 and CCA1 transcript levels. A reduction of these transcript levels could be anticipated in plant lines with increased AtHESP levels. In plants grown in 12/12 h photoperiod, the increase of AtHESP transcript accumulation resulted in a retarded growth phenotype and severe reduction in root length that was attenuated over time, although specific clock-dependent responses were not recorded (). Importantly, the diurnal expression pattern of AtHESP persisted (). Focusing our analysis on the expression levels of TOC1 and CCA1, the diurnal oscillation of both genes retained their normal cycling in AtHESPox lines (). In addition, the expression levels of both genes were also increased.
Figure 6. Diurnal time course of AtHESP, TOC1 and CCA1 expression in A. thaliana overexpression AtHESP-ox plants. (A). Growth and root length retardation phenotypes of AtHESP overexpressing plant lines (AtHESPox) (right panels) compared to wild type plants (left panels) grown for 7, 14 and 21 days in in vitro conditions. (B). AtHESP, TOC1 and CCA1 expression from 3 independent plant lines (AtHESPox14, AtHESPox20and AtHESPox21) with variably up-regulation of AtHESP compared to wild type (wt) plants at 2 time points is shown. Plants were grown under 12 h/12 h light/dark photoperiod. White/black bars at the top indicate light-dark conditions, respectively. Light gray frames indicate subjective dark periods. Relative gene expression was measured with respect to internal housekeeping Ubiquitin gene transcripts. Error bars represent standard error of means (n=3, each consisting of 15 plants). Statistical comparisons within plant lines were performed by Duncan tests (a <0.05). Indicator letters in common denote lack of significant difference. Three independent experimental trials yielded similar results.
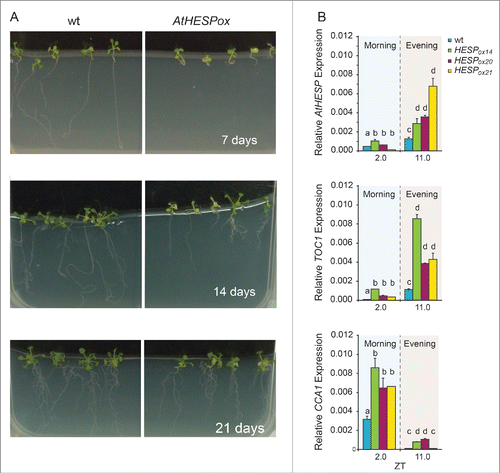
This lack of phase inversion in mRNA accumulation of TOC1 or CCA1, suggests that neither of genes are direct targets of the deadenylase activity of AtHESP.
A better use of absorbed light energy in hesp knock down mutants
Our observation that the AtHESP overexpressing lines had a reversible growth retardation phenotype implied for a negative regulatory role in plant growth. In order to further characterize the hesp mutants, the functioning of the photosynthetic apparatus was studied in comparison to wild-type plants grown under normal growth conditions. To this end, we performed chlorophyll fluorescence measurements Citation61-64 to evaluate whether hesp mutants are affected in PSII photochemistry, and whether AtHESP influences the allocation of absorbed light energy in PSII compared to wild type plants. We estimated the allocation of absorbed light energy to photosynthesis, thermal dissipation and excess excitation, by the energy allocation model of Kramer et al.Citation36 This model allows assessment of excitation energy flux at PSII in 3 different pathways, namely photochemical utilization (ΦPSII), regulated heat dissipation (ΦNPQ, a loss process serving for protection), and non-regulated dissipation (ΦNO, a loss process due to PSII inactivity). The effective quantum yield of photochemical energy conversion in PSII (ΦPSII), was 6.4% less in wild-type plants compared to hesp mutants, indicating that a fraction of absorbed irradiance was not utilized via photochemical reactions in wild type plants, while hesp mutants having a 22% (P < 0.01) less energy dissipated as heat (ΦNPQ), possessed also 1.4% lower (ns different) non-regulated energy dissipated in PSII (ΦNO) than wild-type plants (). Non-photochemical quenching that reflects heat dissipation of excitation energy in the antenna system (NPQ) remained much higher (20.9%, P < 0.01) in wild-type leaves compared to hesp1-1 leaves (). Excitation pressure, 1-qP, was slightly higher (3.5%) in wild-type leaves (), while electron transport rate (ETR) was lower 6.4% (ns different) in wild-type leaves compared to hesp1-1 leaves (). Thus, under normal growth conditions, hesp mutant plants possess not only higher ΦPSII than wild type plants, but also a higher oxidized redox state of QA, which reduced significantly NPQ, and increased the photochemical efficiency, which in turn, increased the ETR.Citation65 These results indicate that hesp mutants may be better protected against oxidative stress.
Discussion
In this work, we demonstrate that AtHESP, an ortholog to the circadian NOC deadenylase in mammals and other organisms, is conserved in plants. The gene encodes for a 3′-end poly(A) exoribonuclease and AtHESP belongs to the EEP superfamily of deadenylases. The gene is regulated by the circadian clock and also influences circadian transcript oscillators, indicating deadenylation and mRNA turnover as a mechanism of regulation of circadian rhythms. In addition, a role for AtHESP in oxidative stress response of plants may be also suggested.
The biochemical characterization of the orthologous NOC from other organisms is still rudimentary. AtHESP shows a high similarity with CNOT6 and CNOT6L deadenylases of the EEP superfamily. Studies in yeast with CCR4 deadenylase (homolog of CNOT6) have shown that poly(A) is the preferred substrate, while poly(U), poly(C), and poly(G) were much more inhibitory than poly(A)Citation66 for degradation activity. Accordingly, our results show that AtHESP prefers poly(A) for degradation. Poly(A) is also the preferred substrate for PARN that belongs to the DEDD superfamily, followed by poly(U) that is degraded less effectively, while poly(C) and poly(G) are very poor substrates.Citation28,67 Further studies on mutagenic analysis of the predicted conserved residues are expected to provide insight into the catalytic mechanism.
The allosteric behavior of AtHESP reported in this work may be understood keeping in mind the key role of the poly(A) tail in mRNA stability. Deadenylases are highly regulated enzymes and their activity is affected by various parameters, such as subcellular localization, posttranslational modifications, cis-acting elements of target mRNAs and trans-acting regulatory factors [for example, see Citation16,68,69]. PARN, which is an extensively studied deadenylase, is indeed an allosterically regulated deadenylase existing in oligomeric structures; its activity is modulated by cis elements, such as 5′-cap, AU-rich and cytoplasmic polyadenylation elements of mRNA, and trans factors, including protein factors, such as the eukaryotic translation initiation factor, the cap-binding complex and cleavage-stimulating factor-50, as well as small molecules, such as nucleotides and nucleoside analogs.Citation68,70 Additionally, PARN exists as a homodimer in dilute solutions, and this is essential for poly(A)-specific activity.Citation71 However, it can also self-associate into active tetramer and high-order oligomers both in vitro and in living cells.Citation72 Our results show that AtHESP is oligomeric and this behavior may represent a regulatory mechanism. Moreover, it has been proposed that deadenylases act in concert and multiple deadenylases can act on the same mRNA, with discrete but overlapping functions.Citation16,73,74 For instance, it has been shown that in mouse cells cytoplasmic deadenylation follows biphasic kinetics; PAN2 deadenylase catalyzes the first phase of poly(A) shortening followed by the activity of CCR4-CAF1 complex.Citation74 Therefore, the diversity of the deadenylases, the complexity of the binding partners, as well as their regulation make up a complicated network to precisely regulate the intracellular mRNA homeostasis.Citation69 The identification of binding partners of AtHESP is expected to contribute to the understanding of its allosteric behavior, while the determination of its distinct domains is anticipated to provide the structural basis for its regulatory requirements.
AtHESP contributes to the control of circadian gene expression and the results of this work suggest deadenylation and mRNA turnover as a mechanism that influences circadian transcript oscillators. The regulation of transcript accumulation of clock genes (TOC1/CCA1/LHY) is significantly perturbed in hesp mutants grown in light/dark cycles. Notably, in Drosophila, the noc knockdown flies have abnormal rhythmic behavior under constant light as well,Citation75 suggesting convergent mechanisms mediating posttranscriptional regulation of circadian rhythm across kingdoms. The rhythmic expression of the gene may be complicated, involving not only circadian regulation but also various systemic and environmental (e.g. nutrition, stress) cues, as has been suggested for the mammalian Noc gene.Citation76 The underlying deadenylation regulatory mechanism through which HESP degrades its substrate remains unknown. We have overexpressed AtHESP and monitored the expression levels of TOC and CCA1 in the presence of an increased level of AtHESP. In these transgenic plant lines we have detected increased levels of TOC and CCA but the cycling pattern of expression was sustained. This lack of phase inversion in mRNA accumulation of TOC1 or CCA1, suggests that neither of genes are direct targets of the deadenylase activity of AtHESP. Thus, AtHESP may affect the circadian rhythm in Arabidopsis by deadenylating one or more unidentified genes. Taking into account the fact that the recent models describing the circadian clock in plants include multiple, 3 or 4, loops of interchangeable interaction, a global, systems biology, approach will be pursued to identify the target mRNAs of AtHESP. Alternatively, overexpressing AtHESP may not be sufficient to disrupt the complexes formed, and as discussed above, AtHESP may act in concert with other partners, including other deadenylases, to fine tune the regulation of the circadian clock. It is also possible that there is a developmental window that is not represented in the time points analyzed in the present study. This is further supported by the severe growth phenotype observed in the overexpressing lines, which was though time-dependent and attenuated over time.
Although deadenylases in plants have not been extensively studied as in other organisms, it is clear that Arabidopsis uses different deadenylases to target distinct mRNAs Citation18,19,77,78 and a role in various developmental and physiological procedures has been designated to the characterized ones. Two PARN genes have been identified in Arabidopsis, one of which, At1g55870 (AtPARN), is a cytoplasmic deadenylase essential for embryogenesis,Citation17 affecting a subset of embryonic transcripts. This is also the case in mammals, where PARN target specific transcripts (reviewed in Reference 68). In addition, 11 CAF1 genes are present in Arabidopsis, some of them linked to stress.Citation18,19,71,77 CAF1 has also been linked to resistance of plants to fungal and bacterial infection, implying important roles in plant-pathogen interactions.Citation19,77,78 The existence of multiple deadenylases may reflect on the different needs for expression of specific mRNAs at different times in cells or tissues, but also on the presence of specific mRNAs in different time periods.Citation16
AtHESP may have a role in plant growth and development as well as in response to stress; the overexpressing plant lines have a severe, albeit reversible retardation phenotype and hesp mutants appear to be better protected against oxidative stress. The reported role of NOC proteins orthologous to AtHESP spans from associations with lipid metabolism and a proadipogenic function to proper wing morphogenesis at the pupal stage in mammals and Drosophila, respectively. It is intriguing to consider that AtHESP may play overlapping roles in the regulation of both the circadian clock and clock output processes, such as response to mechanisms against abiotic and/or biotic stresses Citation79,80 as has been described before for other clock components in plants.Citation81-83 The elucidation of the mechanisms of modulation of this novel deadenylase in the regulation and output of the circadian clock in plants and/or plant development and metabolism, especially under various stress conditions consists an attractive challenge.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Datas.zip
Download Zip (3.3 MB)Acknowledgments
We are grateful to Anders Virtanen (Uppsala University, Sweden) for kindly providing us the pBS (+/-) phagemid for the RNA substrate preparations and the plasmid for PARN expression, Kosmas Haralampidis (University of Athens, Greece) for the pCambia35S-eGFPn vector, Michael Moustakas (Aristotle University of Thessaloniki, Greece) for providing the photosynthesis instrument, Vasiliki T. Skamnaki (University of Thessaly) for help with the gel filtration experiments, George A. Kontopidis (University of Thessaly) and kindly providing the Tricorn Superose 12 10/300 GL column, and Eirini Papanastasi for valuable help in DIG-assays.
References
- Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet 2011; 74:141-73; PMID:21924977; http://dx.doi.org/10.1016/B978-0-12-387690-4.00005-2
- Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol 2009; 60:357-77; PMID:19575587; http://dx.doi.org/10.1146/annurev.arplant.043008.092054
- Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet 2011; 74:175-230; PMID:21924978; http://dx.doi.org/10.1016/B978-0-12-387690-4.00006-4
- Nagel DH, Kay SA. Complexity in the wiring and regulation of plant circadian networks. Curr Biol 2012; 22:R648-57; PMID:22917516; http://dx.doi.org/10.1016/j.cub.2012.07.025
- Brunner M, Schafmeier T. Transcriptional and post-transcriptional regulation of the circadian clock of cyanobacteria and Neurospora. Genes Dev 2006; 20:1061-74; PMID:16651653; http://dx.doi.org/10.1101/gad.1410406
- Henriques R, Mas P. Chromatin remodeling and alternative splicing: pre- and post-transcriptional regulation of the Arabidopsis circadian clock. Semin Cell Dev Biol 2013; 24:399-406; PMID:23499867; http://dx.doi.org/10.1016/j.semcdb.2013.02.009
- van Ooijen G, Millar AJ. Non-transcriptional oscillators in circadian timekeeping. Trends Biochem Sci 2012; 37:484-92; PMID:22917814; http://dx.doi.org/10.1016/j.tibs.2012.07.006
- Staiger D, Green R. RNA-based regulation in the plant circadian clock. Trends Plant Sci 2011; 16:517-23; PMID:21782493; http://dx.doi.org/10.1016/j.tplants.2011.06.002
- So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J 1997; 16:7146-55; PMID:9384591; http://dx.doi.org/10.1093/emboj/16.23.7146
- Woo KC, Ha DC, Lee KH, Kim DY, Kim TD, Kim KT. Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol Cell Biol 2010; 30:197-205; PMID:19858287; http://dx.doi.org/10.1128/MCB.01154-09
- Woo KC, Kim TD, Lee KH, Kim DY, Kim W, Lee KY, Kim KT. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res 2009; 37:26-37; PMID:19010962; http://dx.doi.org/10.1093/nar/gkn893
- Guo J, Cheng P, Yuan H, Liu Y. The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell 2009; 138:1236-46; PMID:19747717; http://dx.doi.org/10.1016/j.cell.2009.06.043
- Lidder P, Gutierrez RA, Salome PA, McClung CR, Green PJ. Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol 2005; 138:2374-85; PMID:16055688; http://dx.doi.org/10.1104/pp.105.060368
- Yakir E, Hilman D, Harir Y, Green RM. Regulation of output from the plant circadian clock. FEBS J 2007; 274:335-45; PMID:17229141; http://dx.doi.org/10.1111/j.1742-4658.2006.05616.x
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell 2009; 136:763-76; PMID:19239894; http://dx.doi.org/10.1016/j.cell.2009.01.019
- Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol 2008; 9:337-44; PMID:18334997; http://dx.doi.org/10.1038/nrm2370
- Reverdatto SV, Dutko JA, Chekanova JA, Hamilton DA, Belostotsky DA. mRNA deadenylation by PARN is essential for embryogenesis in higher plants. Rna 2004; 10:1200-14; PMID:15247430; http://dx.doi.org/10.1261/rna.7540204
- Liang W, Li C, Liu F, Jiang H, Li S, Sun J, Wu X, Li C. The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res 2009; 19:307-16; PMID:19065152; http://dx.doi.org/10.1038/cr.2008.317
- Walley JW, Kelley DR, Nestorova G, Hirschberg DL, Dehesh K. Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol 2010; 152:866-75; PMID:19955262; http://dx.doi.org/10.1104/pp.109.149005
- Dupressoir A, Morel AP, Barbot W, Loireau MP, Corbo L, Heidmann T. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics 2001; 2:9; PMID:11747467; http://dx.doi.org/10.1186/1471-2164-2-9
- Green CB, Besharse JC. Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proc Natl Acad Sci U S A 1996; 93:14884-8; PMID:8962150; http://dx.doi.org/10.1073/pnas.93.25.14884
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 1999; 27:297-300; PMID:9847208; http://dx.doi.org/10.1093/nar/27.1.297
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731-9; PMID:21546353; http://dx.doi.org/10.1093/molbev/msr121
- Cheng Y, Liu WF, Yan YB, Zhou HM. A nonradioactive assay for poly(a)-specific ribonuclease activity by methylene blue colorimetry. Protein Pept Lett 2006; 13:125-8; PMID:16472073; http://dx.doi.org/10.2174/092986606775101580
- Balatsos NA, Anastasakis D, Stathopoulos C. Inhibition of human poly(A)-specific ribonuclease (PARN) by purine nucleotides: kinetic analysis. J Enzyme Inhib Med Chem 2009; 24:516-23; PMID:18763168; http://dx.doi.org/10.1080/14756360802218763
- Greiner-Stoeffele T, Grunow M, Hahn U. A general ribonuclease assay using methylene blue. Anal Biochem 1996; 240:24-8; PMID:8811875; http://dx.doi.org/10.1006/abio.1996.0326
- Åström J, Åström A, Virtanen A. In vitro deadenylation of mammalian mRNA by a HeLa cell 3′ exonuclease. Embo J 1991; 10:3067-71; PMID:1717259
- Martinez J, Ren YG, Thuresson AC, Hellman U, Astrom J, Virtanen A. A 54-kDa fragment of the Poly(A)-specific ribonuclease is an oligomeric, processive, and cap-interacting Poly(A)-specific 3′ exonuclease. J Biol Chem 2000; 275:24222-30; PMID:10801819; http://dx.doi.org/10.1074/jbc.M001705200
- Palfner K, Kneba M, Hiddemann W, Bertram J. Short technical reports. Quantification of ribozyme-mediated RNA cleavage using silver-stained polyacrylamide gels. Biotechniques 1995; 19:926-9; PMID:8747658; http://dx.doi.org/10.1515/bchm3.1995.376.5.289
- Hao ZM, Luo JY, Cheng J, Li L, He D, Wang QY, Yang GX. Intensive inhibition of hTERT expression by a ribozyme induces rapid apoptosis of cancer cells through a telomere length-independent pathway. Cancer Biol Ther 2005; 4:1098-103; PMID:16205109; http://dx.doi.org/10.4161/cbt.4.10.2016
- Berry MJ, Samuel CE. Detection of subnanogram amounts of RNA in polyacrylamide gels in the presence and absence of protein by staining with silver. Anal Biochem 1982; 124:180-4; PMID:6181714; http://dx.doi.org/10.1016/0003-2697(82)90235-4
- Schägger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 1994; 217:220-30; PMID:8203750; http://dx.doi.org/10.1006/abio.1994.1112
- Segel IH. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. Wiley-Interscience; Wiley Classics Library Edition Published 1993 1975:346-85.
- Sperdouli I, Moustakas M. Interaction of proline, sugars, and anthocyanins during photosynthetic acclimation of Arabidopsis thaliana to drought stress. J Plant Physiol 2012; 169:577-85; PMID:22305050; http://dx.doi.org/10.1016/j.jplph.2011.12.015
- Oxborough K, Baker N. Resolving chlorophyll a fluorescence images of photosynthetic efficiency into photochemical and non-photochemical components – calculation of qP and Fv-/Fm-; without measuring Fo. Photosynth Res 1997; 54:135-42; http://dx.doi.org/10.1023/A:1005936823310
- Kramer D, Johnson G, Kiirats O, Edwards G. New Fluorescence Parameters for the Determination of QA Redox State and Excitation Energy Fluxes. Photosynth Res 2004; 79:209-18; PMID:16228395; http://dx.doi.org/10.1023/B:PRES.0000015391.99477.0d
- Schreiber U, Bilger W, Neubauer C. Chlorophyll Fluorescence as a Nonintrusive Indicator for Rapid Assessment of In Vivo Photosynthesis. In: Schulze E-D, Caldwell M, eds. Ecophysiology of Photosynthesis: Springer Berlin Heidelberg, 1995:49-70.
- Gray GR, Savitch LV, Ivanov AG, Huner N. Photosystem II Excitation Pressure and Development of Resistance to Photoinhibition (II. Adjustment of Photosynthetic Capacity in Winter Wheat and Winter Rye). Plant Physiol 1996; 110:61-71; PMID:12226171
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45; PMID:11328886; http://dx.doi.org/10.1093/nar/29.9.e45
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 2003; 339:62-6; PMID:12618301; http://dx.doi.org/10.1016/S0304-3940(02)01423-4
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 2008; 4:e14; PMID:18248097; http://dx.doi.org/10.1371/journal.pgen.0040014
- Priest HD, Filichkin SA, Mockler TC. Cis-regulatory elements in plant cell signaling. Curr Opin Plant Biol 2009; 12:643-9; PMID:19717332; http://dx.doi.org/10.1016/j.pbi.2009.07.016
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001; 293:880-3; PMID:11486091; http://dx.doi.org/10.1126/science.1061320
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 2000; 290:2110-3; PMID:11118138; http://dx.doi.org/10.1126/science.290.5499.2110
- Menkens AE, Cashmore AR. Isolation and characterization of a fourth Arabidopsis thaliana G-box-binding factor, which has similarities to Fos oncoprotein. Proc Natl Acad Sci U S A 1994; 91:2522-6; PMID:8146148; http://dx.doi.org/10.1073/pnas.91.7.2522
- McDonald MJ, Rosbash M, Emery P. Wild-type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol Cell Biol 2001; 21:1207-17; PMID:11158307; http://dx.doi.org/10.1128/MCB.21.4.1207-1217.2001
- Riedl CC, Brader P, Zanzonico PB, Chun YS, Woo Y, Singh P, Carlin S, Wen B, Ling CC, Hricak H, et al. Imaging hypoxia in orthotopic rat liver tumors with iodine 124-labeled iodoazomycin galactopyranoside PET. Radiology 2008; 248:561-70; PMID:18641253; http://dx.doi.org/10.1148/radiol.2482071421
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 1997; 9:491-507; PMID:9144958; http://dx.doi.org/10.1105/tpc.9.4.491
- Chen J, Chiang YC, Denis CL. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J 2002; 21:1414-26; PMID:11889047; http://dx.doi.org/10.1093/emboj/21.6.1414
- Dupressoir A, Barbot W, Loireau MP, Heidmann T. Characterization of a mammalian gene related to the yeast CCR4 general transcription factor and revealed by transposon insertion. J Biol Chem 1999; 274:31068-75; PMID:10521507; http://dx.doi.org/10.1074/jbc.274.43.31068
- Wang Y, Osterbur DL, Megaw PL, Tosini G, Fukuhara C, Green CB, Besharse JC. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev Biol 2001; 1:9; PMID:11394964; http://dx.doi.org/10.1186/1471-213X-1-9
- Draper MP, Salvadore C, Denis CL. Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol Cell Biol 1995; 15:3487-95; PMID:7791755; http://dx.doi.org/10.1128/MCB.15.7.3487
- Wang H, Morita M, Yang X, Suzuki T, Yang W, Wang J, Ito K, Wang Q, Zhao C, Bartlam M, et al. Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J 2010; 29:2566-76; PMID:20628353; http://dx.doi.org/10.1038/emboj.2010.152
- Mittal S, Aslam A, Doidge R, Medica R, Winkler GS. The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4-Not complex contribute to the prevention of cell death and senescence. Molecular biology of the cell 2011; 22:748-58; PMID:21233283; http://dx.doi.org/10.1091/mbc.E10-11-0898
- Balatsos NA, Vlachakis D, Maragozidis P, Manta S, Anastasakis D, Kyritsis A, Vlassi M, Komiotis D, Stathopoulos C. Competitive inhibition of human poly(A)-specific ribonuclease (PARN) by synthetic fluoro-pyranosyl nucleosides. Biochemistry 2009; 48:6044-51; PMID:19472977; http://dx.doi.org/10.1021/bi900236k
- Åström J, Åström A, Virtanen A. Properties of a HeLa cell 3′ exonuclease specific for degrading poly(A) tails of mammalian mRNA. J Biol Chem 1992; 267:18154-9
- Ding Z, Doyle MR, Amasino RM, Davis SJ. A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation. Genetics 2007; 176:1501-10; PMID:17483414; http://dx.doi.org/10.1534/genetics.107.072769
- Ito S, Nakamichi N, Nakamura Y, Niwa Y, Kato T, Murakami M, Kita M, Mizoguchi T, Niinuma K, Yamashino T, et al. Genetic linkages between circadian clock-associated components and phytochrome-dependent red light signal transduction in Arabidopsis thaliana. Plant Cell Physiol 2007; 48:971-83; PMID:17519251; http://dx.doi.org/10.1093/pcp/pcm063
- Niwa Y, Ito S, Nakamichi N, Mizoguchi T, Niinuma K, Yamashino T, Mizuno T. Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol 2007; 48:925-37; PMID:17540692; http://dx.doi.org/10.1093/pcp/pcm067
- Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G, Ohkura N, Azama T, Mesaki M, Yukimasa S, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem 2003; 278:41519-27; PMID:12865428; http://dx.doi.org/10.1074/jbc.M304564200
- Baker NR. Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol 2008; 59:89-113; PMID:18444897; http://dx.doi.org/10.1146/annurev.arplant.59.032607.092759
- Maxwell K, Johnson GN. Chlorophyll fluorescence–a practical guide. J Exp Bot 2000; 51:659-68; PMID:10938857; http://dx.doi.org/10.1093/jexbot/51.345.659
- Murchie EH, Lawson T. Chlorophyll fluorescence analysis: a guide to good practice and understanding some new applications. J Exp Bot 2013; 64:3983-98; PMID:23913954; http://dx.doi.org/10.1093/jxb/ert208
- Papageorgiou G. Fluorescence of Photosynthetic Pigments in Vitro and in Vivo. In: Papageorgiou G, Govindjee, eds. Chlorophyll a Fluorescence: Springer Netherlands, 2004:43-63.
- Tu W, Li Y, Zhang Y, Zhang L, Liu H, Liu C, Yang C. Diminished photoinhibition is involved in high photosynthetic capacities in spring ephemeral Berteroa incana under strong light conditions. J Plant Physiol 2012; 169:1463-70; PMID:22854181; http://dx.doi.org/10.1016/j.jplph.2012.05.027
- Viswanathan P, Chen J, Chiang YC, Denis CL. Identification of multiple RNA features that influence CCR4 deadenylation activity. J Biol Chem 2003; 278:14949-55; PMID:12590136; http://dx.doi.org/10.1074/jbc.M211794200
- Korner CG, Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J Biol Chem 1997; 272:10448-56; PMID:9099687; http://dx.doi.org/10.1074/jbc.272.1.96
- Balatsos NA, Maragozidis P, Anastasakis D, Stathopoulos C. Modulation of poly(A)-specific ribonuclease (PARN): current knowledge and perspectives. Curr Med Chem 2012; 19:4838-49; PMID:22834816; http://dx.doi.org/10.2174/092986712803341539
- Yan YB. Deadenylation: enzymes, regulation, and functional implications. Wiley Interdiscip Rev RNA 2014; 5:421-43; PMID:24523229; http://dx.doi.org/10.1002/wrna.1221
- Virtanen A, Henriksson N, Nilsson P, Nissbeck M. Poly(A)-specific ribonuclease (PARN): an allosterically regulated, processive and mRNA cap-interacting deadenylase. Crit Rev Biochem Mol Biol 2013; 48:192-209; PMID:23496118; http://dx.doi.org/10.3109/10409238.2013.771132
- Wu M, Reuter M, Lilie H, Liu Y, Wahle E, Song H. Structural insight into poly(A) binding and catalytic mechanism of human PARN. Embo J 2005; 24:4082-93; PMID:16281054; http://dx.doi.org/10.1038/sj.emboj.7600869
- He GJ, Yan YB. Self-association of poly(A)-specific ribonuclease (PARN) triggered by the R3H domain. Biochimica et biophysica acta 2014; 1844:2077-85; PMID:25239613; http://dx.doi.org/10.1016/j.bbapap.2014.09.010
- Zhang LN, Yan YB. Depletion of poly(A)-specific ribonuclease (PARN) inhibits proliferation of human gastric cancer cells by blocking cell cycle progression. Biochim Biophys Acta 2015; 1853:522-34; PMID:25499764; http://dx.doi.org/10.1016/j.bbamcr.2014.12.004
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 2005; 12:1054-63; PMID:16284618; http://dx.doi.org/10.1038/nsmb1016
- Nagoshi E, Sugino K, Kula E, Okazaki E, Tachibana T, Nelson S, Rosbash M. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci 2010; 13:60-8; PMID:19966839; http://dx.doi.org/10.1038/nn.2451
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 2007; 5:e34; PMID:17298173; http://dx.doi.org/10.1371/journal.pbio.0050034
- Walley JW, Kelley DR, Savchenko T, Dehesh K. Investigating the function of CAF1 deadenylases during plant stress responses. Plant Signal Behav 2010; 5:802-5; PMID:20421740; http://dx.doi.org/10.4161/psb.5.7.11578
- Sarowar S, Oh HW, Cho HS, Baek KH, Seong ES, Joung YH, Choi GJ, Lee S, Choi D. Capsicum annuum CCR4-associated factor CaCAF1 is necessary for plant development and defence response. Plant J 2007; 51:792-802; PMID:17587232; http://dx.doi.org/10.1111/j.1365-313X.2007.03174.x
- Doherty CJ, Kay SA. Circadian Control of Global Gene Expression Patterns. Annual review of genetics 2010; 44:419-44; PMID:20809800; http://dx.doi.org/10.1146/annurev-genet-102209-163432
- Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JHM, Dijkwel PP. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci 2012; 109:17129-34; PMID:23027948; http://dx.doi.org/10.1073/pnas.1209148109
- Farre EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 2005; 15:47-54; PMID:15649364; http://dx.doi.org/10.1016/j.cub.2004.12.067
- Liu T, Carlsson J, Takeuchi T, Newton L, Farré EM. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J 2013; 76:101-14; PMID:23808423; http://dx.doi.org/10.1111/tpj.12276
- Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 2009; 50:447-62; PMID:19131357; http://dx.doi.org/10.1093/pcp/pcp004

