ABSTRACT
ADAR1 is an enzyme that belongs to the Adenosine Deaminases Acting on RNA (ADARs) family. These enzymes deaminate adenosines to inosines (RNA editing A-to-I) within double-stranded RNA regions in transcripts. Since inosines are recognized as guanosines by the cellular machinery, RNA editing mediated by ADARs can either lead to the formation of an altered protein (recoding) or affect different aspects of RNA metabolism.
Recently, a proteomic analysis led to the identification of novel ADAR1-associated factors and found that a good fraction of them is shared with the Long Interspersed Element 1 (LINE-1 or L1) ribonucleoparticles (RNPs). This evidence suggested a possible role of ADAR1 in regulating the L1 life cycle. By taking advantage of the use of cell culture retrotransposition assays, a novel function of this deaminase as an inhibitor of L1 retrotransposition was demonstrated. These results pave the way toward a better comprehension of the mechanisms of restriction of retrotransposons.
Introduction
Retrotransposons, are the predominant class of Transposable Elements (TEs) in most mammalian genomes and can be subdivided into those sequences that contain the long-terminal repeats (LTR) and those that do not (non-LTR).Citation1-2 The Long INterspersed Element-1 (LINE-1s or L1s) is an autonomous non-LTR retrotransposon that continues to generate both intra-and inter-individual genetic variations in the human population.Citation1
A typical retrotransposition-competent human LINE-1 element is ∼6 kb in length and contains a 5′ untranslated region (UTR), open reading frames (ORFs) and a 3′ UTR.Citation3-4
The promoter region of these elements is within the 5′ UTR, driving the transcription of a bicistronic L1 RNA, which contains 2 non-overlapping ORFs (ORF1 and ORF2).Citation5
ORF1p is a ∼40 kDa polypeptide with nucleic acid chaperone activity,Citation6-7 and ORF2p is a ∼150 kDa protein with endonuclease and reverse transcriptase functions.Citation8-10 Upon translation the ORF1p and ORF2p proteins bind their own L1 RNA (cis-preference) forming a ribonucleoprotein (RNP) complex in the cytoplasm.Citation11-12 Upon translocation of the RNP to the nucleus, it may generate a new chromosomal insertion via a target primed reverse transcription mechanism (TPRT).Citation13 L1 can also function in trans to mobilize the non-autonomous Short Interspersed Elements (SINEs).
Even though more than 500 000 copies of L1 exist in the human genome, most L1s are inactive due to point mutations, rearrangements, or truncations, with only 80–100 elements potentially active in any individual.Citation14-15
The uncontrolled retrotransposition of LINE-1 can be deleterious for the host genome thereby the cell has evolved several mechanisms of defense against these endogenous parasites. The repression of retrotransposons occurs both at the transcriptional and post-transcriptional levels.Citation16-17
The major control occurs by limiting the expression of L1 through histone modification and DNA methylation.Citation18-25
Furthermore, several cellular trans-acting restriction factors regulate the L1 life cycle with different mechanism.Citation16-17 The list of these factors is growing fast and interestingly many of these proteins are involved in nucleic acid metabolism, and some are induced by type I interferons.
During evolution, the cell has developed different mechanisms of defense to protect against the danger of endogenous and exogenous parasites, thus it is not surprising that many of the known anti-retrotransposon restriction factors are also anti-retroviral. Of note, different enzymes belonging to the family of the APOBEC3 cytidine deaminase were reported to restrict LINE-1 retrotransposition. In particular, APOBEC3A can inhibit retrotransposition through the deamination of the single-stranded DNAs that are exposed transiently during the LINE-1 TPRT process,Citation26 whereas APOBEC3B, APOBEC3C, and APOBEC3DE seem to inhibit retrotransposition by diverse deamination-independent mechanisms,Citation27-29 thus showing the complexity of the restriction processes. Moreover, we recently provided evidence showing that another deaminase, ADAR1, is a suppressor of LINE-1 retrotransposition.Citation30
ADAR1 is a member of the Adenosine Deaminases Acting on RNA (ADARs) family and catalyzes the conversion of adenosine to inosine (A-to-I) within double-stranded RNAs.Citation31-32 There are 2 different ADAR1 isoforms generated by the use of alternative promoters and alternative splicing.Citation33-35 The short form of ADAR1 (p110) is constitutively expressed and localizes mainly to the nucleus, whereas the long form of ADAR1 (p150) is interferon-inducible and shuttles between the nucleus and cytoplasm.Citation31 ADAR1 can edit both cellular and viral RNAs and since inosines are recognized as guanosines by the cellular machinery, RNA editing mediated by ADARs can lead to the formation of an altered protein if editing occurs within the coding sequence of mRNAs.Citation31,Citation36 However, recently it has been clearly demonstrated that most of the RNA editing occurs within non-coding regions, most particularly Alu elements, affecting different aspects of the RNA metabolism, such as RNA stability, translation, splicing, and interaction with specific protein factors.Citation36 Moreover, RNA editing is deregulated in a variety of human diseases.Citation32,Citation37-41 Therefore, ADAR1 has a deep impact on gene expression regulation. Moreover, ADAR1 plays a role as suppressor of interferon (IFN) signaling.Citation42-43 Recently, Mannion and colleagues proposed that ADAR1 editing may mark endogenous dsRNAs as “self” to distinguish them from exogenous, mostly viral, “non self” dsRNAs thus avoiding the induction of an aberrant type I IFN response.Citation44 This hypothesis was further confirmed by othersCitation45-47
ADAR1 is an inhibitor of LINE-1 retrotransposition
Our study started with the affinity purification of the ADAR1 RNP complexes followed by mass spectrometry analysis of 293T cells expressing HIV-1 with the initial goal of identifying the ADAR1-interacting factors that could contribute to the proviral activity of the deaminase previously reported.Citation30,Citation48-50 This analysis led to the identification of 14 non-ribosomal ADAR1-associated factors, among which a good fraction, such as PABPC1, hnRNP L, HSPA1A, nucleolin and TOP1 were previously reported as L1 RNP-associated factors.Citation51-53 Moreover, 3 of these proteins (nucleolin, hnRNP L and PABPC1) were shown to affect L1 retrotransposition.Citation51,Citation54-55 This result prompted us to test whether ADAR1 too is involved in the L1 life cycle. First, we confirmed by co-immunoprecipitation experiments the interaction between ADAR1 and the L1 RNP-associated proteins identified by mass spectrometry. Moreover, we found that these interactions occur also in the absence of HIV-1 expression, thus suggesting that we identified general interactors of the deaminase, most of which are novel. Furthermore, we tested whether ADAR1 is a regulator of L1 retrotransposition by taking advantage of the use of different and widely used cell culture retrotransposition assays.Citation56-58
By using these assays in HeLa cells silenced for ADAR1 expression, we observed an increase of LINE-1 retrotransposition. We further extended our analysis and confirmed that overexpression of ADAR1 decreases L1 retrotransposition.Citation30 Overall, these results suggest a novel function for ADAR1 as a general repressor of retrotransposition.
Possible mechanism for ADAR1 anti-retrotransposon activity
What could be the mechanism that drives the inhibition of L1 retrotransposition mediated by ADAR1?
a) RNA editing model
It was previously suggested that L1 RNAs harbor some double-stranded (ds) RNA-binding elements for Microprocessor,Citation59 thus the most logical answer to this question is that ADAR1 upon binding to L1 dsRNA, catalyzes the conversion of adenosines to inosines, thus potentially either altering the folding of some dsRNA elements or mutating sequences that are critical for retrotransposition activity ().
Figure 1. Possible models for how ADAR1 inhibits LINE-1 retrotransposition. (A) L1 RNAs are edited by ADAR1 causing nucleotide changes in the ORF sequence or altering the dsRNA elements harbored within L1 RNAs. (B) ADAR1 by binding the basal L1 RNP complex and/or its associated proteins impairs its functionality. (C) ADAR1 sequesters L1 RNP complexes in the stress granules. (D) ADAR1 binds L1 DNA/RNA hybrids during the RT step of the L1 life cycle and either impairs the formation of these hybrids or edits the DNA.
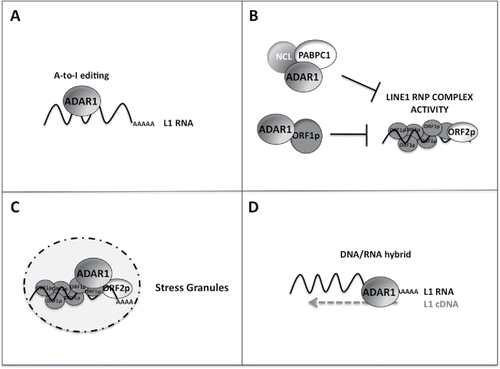
We tested this hypothesis by using a deletion mutant of ADAR1 that lacks the deaminase domain, and showed that this protein represses L1 retrotransposition to a similar extent as the wild-type protein.Citation30 Moreover, we did not find A-to-I editing events in the ectopically expressed L1 RNAs isolated from cells over-expressing ADAR1 (almost half of the full-length 6 kb analyzed).Citation30 All together, these results suggest that ADAR1 inhibits retrotransposition by a mechanism that is RNA-editing independent, even though a more detailed analysis is required to fully address this issue. In fact, we cannot exclude that editing is indeed occurring at a very low frequency below the threshold of detection of our assay (3–4%), although not sufficiently to affect the retrotransposition process. Moreover, we cannot rule out that editing is occurring in the L1 RNA regions not sequenced.
To further complicate this issue, recently, by using inosine-specific cyanoethylation combined with Sanger sequencing (ICE method) and deep sequencing (ICE-seq), several A-to-I editing events were identified in the human brain transcriptome, and some of them lie in the 3′UTRs of transcripts (i.e. TAF1, ACBD7, GPLD1, TAF1, NBPF11 mRNAs) containing dsRNA structures formed by LINE sequences (partial, not full length) repeated in tandem and in inverse orientation.Citation60 Therefore, LINE-1 sequences embedded in other transcripts and forming dsRNA structures can be targets of ADAR enzymes.
b) Restriction of L1-RNP complex
In our study, we also reported that ADAR1 binds to the basal L1 RNP complex, in particular ORF1p and the L1 RNA.Citation30 The correct assembly of the L1 RNP complex is pivotal for retrotransposon activity, thus it is plausible that ADAR1 binding to the L1 RNP complex might directly or indirectly interfere with its activity resulting in the inhibition of retrotransposition (). We first assayed whether ADAR1 binding to the L1 RNP complex might affect the intracellular accumulation of L1 RNA and ORF1p components, as shown for other L1 restriction factors.Citation53,Citation61-64
We tested this hypothesis by measuring the levels of both ORF1p (by Western Blot analysis) and L1 RNA (by RT-qPCR) ectopically expressed from a retrotransposition cassette in HeLa cells silenced for ADAR1 expression, but we did not find any significant alteration in their levels compared with the controls.Citation30 This result indicates that in our experimental setting the ADAR1 inhibition of L1 retrotransposition is not caused by a decreased stability/accumulation of the L1 RNP components. Nevertheless, silencing of ADAR1 expression in HeLa cells causes an increase of the endogenous L1 transcripts (measured by RT-qPCR, Orecchini et al. unpublished). We don't know the reason for this discrepancy and additional experiments are required to explain it.
Furthermore, as mentioned above, we have shown the interaction between ADAR1 and some L1 RNP-associated proteins.Citation30 In particular, PABPC1 and nucleolin proteins were previously reported to exert a positive effect on L1 retrotransposition.Citation54-55 In fact, PABPC1 is critical for L1 RNP formation, and alteration of its intracellular level affects retrotransposition and subcellular localization of ORF1p.Citation54 Moreover, nucleolin likely acts as an IRES trans-acting factor to stimulate ORF2 translation of murine L1 RNA.Citation55
Therefore, ADAR1 by interacting with these proteins and other L1 RNP-associated factors may affect their stimulatory activity thus impairing L1 retrotransposition ().
Finally, we have preliminary results showing that ADAR1 may also impair Alu retrotransposition (Orecchini et al., unpublished). In particular, in HeLa cells silenced for ADAR1 expression and co-transfected with an Alu retrotransposition cassette (pAlu-Neotet)Citation65 together with an ORF2p expression vector (pORF2NoNeo), an increase in Alu retrotransposition was observed. This result suggests that ADAR1 regulates different classes of retrotransposons. Since in this assay the Alu retrotransposition can be achieved only through the overexpression of the LINE-1 ORF2p protein, it would be of great interest to test whether both the inhibition of L1 and Alu retrotransposition mediated by ADAR1 occur through the disruption of ORF2p activity or by reducing its level as previously shown for SAMHD1.Citation66
c) Stress Granules as a site for L1 RNP sequestration by ADAR1
It has been previously reported that the L1 RNA, ORF1p and ORF2p proteins accumulate in stress granules (SGs) and in the nucleoli of a small percentage of cells.Citation67-69 SGs are assemblies of untranslating messenger ribonucleoproteins (mRNPs) that form from mRNAs stalled at translation initiation, and their formation modulates the stress response, viral infection, and signaling pathways.Citation70
Of note, ADAR1 p110 isoform is almost exclusively a nuclear/nucleolar protein, while ADAR1 p150 is a shuttling protein and accumulates in the cytoplasm and under particular stress localizes in stress granules (SGs).Citation71 The Zα domain of ADAR1 p150 is required for such specific subcellular localization.Citation71 Moreover, we identified stress granule-associated proteins G3BP2 and PABPC1 as novel interactors of ADAR1.Citation30
Therefore, it is conceivable that SGs and the nucleoli are the subcellular compartments where the interaction between ADAR1 and the L1 RNPs may take place. To address this issue, we performed immunofluorescence experiments in 293T cells transfected with a retrotransposition cassette containing the full length L1 sequence with the ORF1p fused to a T7 tag (pES2TE1; 69). We have preliminary results showing that the endogenous ADAR1 co-localizes with ORF1p in cytoplasmic granules; this localization is even more evident when cells are treated with an inducer of SGs, such as sodium arsenite (Orecchini et al. unpublished).
Interestingly, other L1 RNP-associated factors localize in SGs, such as MOV10, ZAP, PABPC1 and APOBEC3 proteins.Citation53,Citation61,Citation62,Citation67,Citation72 Notably, Hu and collaborators proposed a novel mechanism whereby SAMHD1 enhances assembly of cytoplasmic stress granules that then sequester L1 RNPs and prevent their retrotransposition.Citation73
Therefore, we cannot exclude that ADAR1, in particular the p150 isoform, may suppress L1 retrotransposition through a similar mechanism (). In any event, co-localization of ADAR1 p150 with the ORF1p protein suggests that this isoform may be critical for L1 restriction.
d) DNA/RNA hybrids
Finally, it has been recently demonstrated that ADARs can deaminate 2′-deoxyadenosines in the DNA strands of DNA/RNA hybrids in vitro, thus expanding the possible biologic functions of ADARs.Citation74 The RNA-DNA hybrid is an essential intermediate of reverse transcription during the retrotransposition process of LINE-1, thus it can be envisioned that ADAR1, by simply binding such RNA-DNA hybrids and/or by mutating the L1 DNA sequence may affect retrotransposition (). Based on the results described above using the mutant of ADAR1 lacking the deaminase domain we suppose that the first hypothesis is more plausible.
Aicardi-Goutières syndrome
Aicardi-Goutières Syndrome (AGS) is an inflammatory encephalopathy that exhibits a neurologic dysfunction characterized by increased production of type I interferon (IFN).Citation75
AGS can be caused by mutations in any of 7 genes (TREX1, RNaseH2A, RNaseH2B, RNaseH2C, SAMHD1, ADAR1 and IFIH1) that carry out diverse functions of intracellular nucleic acid metabolism and sensing. A deficiency of these proteins is thought to result in the accumulation of self-derived nucleic acid species that are recognized as danger signals by sensors of the innate immune system, triggering the pathogenic type 1 interferon (IFN) response.Citation75
The source of endogenous nucleic acids that are hypothesized to induce such a response remains uncertain, but may relate to retroelements.
Indeed, the products of 6 out the 7 AGS-related genes can be placed in a common pathway of metabolism of retroelements. In TREX1-deficient cells, type I IFN activation has been attributed to an increased amount of reverse transcribed DNA derived from endogenous retroelements.Citation76 Furthermore, it has been recently shown that TREX1 inhibits L1 retrotransposition by depleting ORF1p protein and AGS-related TREX1 mutants are deficient in this activity.Citation64 Moreover, SAMHD1 has been demonstrated to be a potent inhibitor of L1 activity and importantly it was found that AGS-related SAMHD1 mutants are defective in L1 retrotransposition inhibition.Citation66 It has been suggested that the RNaseH2 may be involved in the suppression of endogenous retroelements,Citation77 and we recently demonstrated that ADAR1 is an inhibitor of LINE-1 retrotransposition.Citation30 In addition, it has been reported that L1 activity is a potential inducer of interferon expression and autoimmune disorders,Citation77-79 thus providing a link between the dysregulation of L1 retrotransposition and the pathogenesis of AGS.
Therefore, we cannot exclude that in AGS patients containing mutations in ADAR1 (AGS6; 80), an increased level of L1 transcripts could trigger an aberrant IFN activation. However, Mannion and collaboratorsCitation44 analyzed repetitive element transcript levels by RNA-Seq in Adar1−/− embryos and no substantial differences were found compared with the control besides an increased expression of individual members of ERV and IAP families in the mutant mice. This result is in disagreement with our results showing an increase of L1 transcripts in HeLa cells silenced for ADAR1 expression, but it may suggest a general role of this enzyme in the metabolism of different retroelements. Therefore, it would be of paramount importance to assay the transcript level of all the different classes of retroelements (with particular attention to the HERV) in AGS6 patients to determine whether their expression is altered in that specific context.
Concluding remarks
Recent studies have provided a deeper knowledge of the different mechanisms causing the restriction of L1 retrotransposition. We found that ADAR1 is among the protein factors that inhibit retrotransposition, probably at the post-transcriptional level. Future investigations are required to shed light on the mechanism through which ADAR1 inhibits retrotransposition and to elucidate whether this enzyme is active against the mobilization of other retroelements.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Dr Moran JV at the University of Michigan
Medical School for pES2TE1 cassettes. The authors also thank John Goodier, Silvia Anna Ciafrè and Silvia Galardi for critical reading of the manuscript.
References
- Elbarbary RA, Lucas BA, Maquat LE. Retrotransposons as regulators of gene expression. Science 2016; (6274):aac7247; PMID: 26912865; https://doi.org/10.1126/science.aac7247
- Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran JV. The Influence of LINE-1 and SINE Retrotransposons on mammalian genomes. Microbiol Spectr 2015; 3(2):MDNA3-0061-2014; PMID:26104698; https://doi.org/10.1128/microbiolspec.MDNA3-0061-2014
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH, Jr. Isolation of an active human transposable element. Science 1991; 254:1805-08; PMID: 1662412; https://doi.org/10.1126/science.1662412
- Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O'Hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L. Origin of the human L1 elements: Proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1987; 1(2):113-25; PMID: 3692483; https://doi.org/10.1016/0888-7543(87)90003-6
- Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol 1990; 10(12):6718-29; PMID: 1701022; https://doi.org/10.1128/MCB.10.12.6718
- Holmes SE, Singer MF, Swergold GD. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J Biol Chem 1992; 267(28):19765-8; PMID: 1328181.
- Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J 1996; 15(3):630-39; PMID: 8599946.
- Ergun S, Buschmann C, Heukeshoven J, Dammann K, Schnieders F, Lauke H, Chalajour F, Kilic N, Strätling WH, Schumann GG. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J Biol Chem 2004; 279(26):27753-63; PMID: 15056671; https://doi.org/10.1074/jbc.M312985200
- Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science 1991; 20; 254(5039):1808-10; PMID: 1722352; https://doi.org/10.1126/science.1722352
- Feng Q, Moran JV, Kazazian HH Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 1996; 87(5):905-16; PMID: 8945517; https://doi.org/10.1016/S0092-8674(00)81997-2
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: Cis preference versus trans complementation. Mol Cell Biol 2001; 21(4):1429-39; PMID: 11158327; https://doi.org/10.1128/MCB.21.4.1429-1439.2001
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet 2000; 24(4):363-67; PMID: 10742098; https://doi.org/10.1038/74184
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72(4):595-605; PMID: 7679954; https://doi.org/10.1016/0092-8674(93)90078-5
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A 2003 Apr 29; 100(9):5280-5; PMID: 12682288; https://doi.org/10.1073/pnas.0831042100
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 retrotransposition activity in human genomes. Cell 2010; 141(7):1159-70; PMID: 20602998; https://doi.org/10.1016/j.cell.2010.05.021
- Goodier JL. Restricting retrotransposons: A review. Mob DNA 2016 Aug 11; 7:16; PMID: 27525044; https://doi.org/10.1186/s13100-016-0070-z
- Pizarro JG, Cristofari G. Post-Transcriptional Control of LINE-1 Retrotransposition by cellular host factors in somatic cells. Front Cell Dev Biol 2016; 4:14; PMID: 27014690; https://doi.org/10.3389/fcell.2016.00014
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 1997; 13(8):335-40; PMID: 9260521; https://doi.org/10.1016/S0168-9525(97)01181-5
- Bestor TH, Bourc'his D. Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb Symp Quant Biol 2004; 69:381-87; PMID: 16117671; https://doi.org/10.1101/sqb.2004.69.381
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet 2011; 13(1):7-13; PMID:22083101; https://doi.org/10.1038/nrg3080
- Kondo Y, Issa JP. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J Biol Chem 2003; 278(30):27658-62; PMID: 12724318; https://doi.org/10.1074/jbc.M304072200
- Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 2005; 24(4):800-12; PMID: 15678104; https://doi.org/10.1038/sj.emboj.7600545
- Hunter RG, Murakami G, Dewell S, Seligsohn M, Baker ME, Datson NA, McEwen BS, Pfaff DW. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc Natl Acad Sci U S A 2012; 109(43):17657-62; PMID: 23043114; https://doi.org/10.1073/pnas.1215810109
- Bulut-Karslioglu A, De La Rosa-Velázquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galán C, Winter GE, Engist B, Gerle B, et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell. 2014; 55(2):277-90; PMID: 24981170; https://doi.org/10.1016/j.molcel.2014.05.029
- Di Giacomo M, Comazzetto S, Sampath SC, Sampath SC, O'Carroll D. G9a co-suppresses LINE1 elements in spermatogonia. Epigenetics Chromatin 2014; 7:24; PMID: 25276231; https://doi.org/10.1186/1756-8935-7-24
- Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. Elife 2014; 3:e02008; PMID: 24843014; https://doi.org/10.7554/eLife.02008
- Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J Biol Chem 2011 Oct 21; 286(42):36427-37; PMID: 21878639; https://doi.org/10.1074/jbc.M111.251058
- Horn AV, Klawitter S, Held U, Berger A, Vasudevan AA, Bock A, Hofmann H, Hanschmann KM, Trösemeier JH, Flory E, et al. Human LINE-1 restriction by APOBEC3C is deaminase independent and mediated by an ORF1p interaction that affects LINE reverse transcriptase activity. Nucleic Acids Res 2014 Jan; 42(1):396-416; PMID: 24101588; https://doi.org/10.1093/nar/gkt898
- Liang W, Xu J, Yuan W, Song X, Zhang J, Wei W, Yu XF, Yang Y. APOBEC3DE Inhibits LINE-1 Retrotransposition by interacting with ORF1p and influencing LINE reverse transcriptase activity. PLoS One 2016; 11(7):e0157220; PMID: 27428332; https://doi.org/10.1371/journal.pone.0157220
- Orecchini E, Doria M, Antonioni A, Galardi S, Ciafrè SA, Frassinelli L, Mancone C, Montaldo C, Tripodi M, Michienzi A. ADAR1 restricts LINE-1 retrotransposition. Nucleic Acids Res 2017; 45(1):155-68; PMID: 27658966; https://doi.org/10.1093/nar/gkw834
- George CX, John L, Samuel CE. An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1). J Interferon Cytokine Res 2014; 34(6):437-46; PMID: 24905200; https://doi.org/10.1089/jir.2014.0001
- Song C, Sakurai M, Shiromoto Y, Nishikura K. Functions of the RNA editing enzyme ADAR1 and their relevance to human diseases. Genes (Basel) 2016; 7(12):pii: E129; PMID:27999332; https://doi.org/10.3390/genes7120129
- Patterson, JB, Samuel, CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol Cell Biol 1995; 15(10):5376-88; PMID: 7565688; https://doi.org/10.1128/MCB.15.10.5376
- George CX, Samuel CE. Characterization of the 5′-flanking region of the human RNA-specific adenosine deaminase ADAR1 gene and identification of an interferon-inducible ADAR1 promoter. Gene 1999; 229(1-2):203-13; PMID: 10095120; https://doi.org/10.1016/S0378-1119(99)00017-7
- George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci U S A 1999; 96(8):4621-26; PMID: 10200312; https://doi.org/10.1073/pnas.96.8.4621
- Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol 2016; 17(2):83-96; PMID: 26648264; https://doi.org/10.1038/nrm.2015.4
- Yamashita T, Akamatsu M, Kwak S. Altered intracellular milieu of ADAR2-Deficient motor neurons in amyotrophic lateral sclerosis. Genes (Basel) 2017; 8(2):pii: E60; PMID:28208729; https://doi.org/10.3390/genes8020060
- Silberberg G, Lundin D, Navon R, Öhman M. Deregulation of the A-to-I RNA editing mechanism in psychiatric disorders. Hum Mol Genet 2012; 21(2):311-21; PMID: 21984433; https://doi.org/10.1093/hmg/ddr461
- Samuel CE. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology 2011; 411(2):180-93; PMID: 21211811; https://doi.org/10.1016/j.virol.2010.12.004
- Zipeto MA, Jiang Q, Melese E, Jamieson CH. RNA rewriting, recoding, and rewiring in human disease. Trends Mol Med 2015; 21(9):549-59; PMID: 26259769; https://doi.org/10.1016/j.molmed.2015.07.001
- Rayon-Estrada V, Papavasiliou FN, Harjanto D. RNA editing dynamically rewrites the cancer code. Trends Cancer 2015; 1(4):211-12; PMID: 27695712; https://doi.org/10.1016/j.trecan.2015.10.008
- Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol 2009; 10(1):109-15; PMID: 19060901; https://doi.org/10.1038/ni.1680
- Li Z, Okonski KM, Samuel CE. Adenosine deaminase acting on RNA 1 (ADAR1) suppresses the induction of interferon by measles virus. J Virol 2012; 86(7):3787-94; PMID: 22278222; https://doi.org/10.1128/JVI.06307-11
- Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellåker C, Vesely C, Ponting CP, McLaughlin PJ, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep 2014; 9(4):1482-94; PMID: 25456137; https://doi.org/10.1016/j.celrep.2014.10.041
- George CX, Ramaswami G, Li JB, Samuel CE. Editing of cellular Self-RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. J Biol Chem 2016; 291(12):6158-68; PMID: 26817845; https://doi.org/10.1074/jbc.M115.709014
- Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 2015; 349(6252):1115-20; PMID: 26275108; https://doi.org/10.1126/science.aac7049
- Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. Isoforms of RNA-Editing enzyme ADAR1 independently control nucleic acid Sensor MDA5-Driven autoimmunity and multi-organ development. Immunity 2015; 43(5):933-44; PMID: 26588779; https://doi.org/10.1016/j.immuni.2015.11.001
- Doria M, Neri F, Gallo A, Farace MG, Michienzi A. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res 2009; 37(17):5848-58; PMID: 19651874; https://doi.org/10.1093/nar/gkp604
- Doria M, Tomaselli S, Neri F, Ciafrè SA, Farace MG, Michienzi A, Gallo A. ADAR2 editing enzyme is a novel human immunodeficiency virus-1 proviral factor. J Gen Virol 2011; 92(Pt 5):1228-32; PMID: 21289159; https://doi.org/10.1099/vir.0.028043-0
- Orecchini E, Federico M, Doria M, Arenaccio C, Giuliani E, Ciafrè SA, Michienzi A. The ADAR1 editing enzyme is encapsidated into HIV-1 virions. Virology 2015; 485:475-80; PMID: 26363218; https://doi.org/10.1016/j.virol.2015.07.027
- Goodier JL, Cheung LE, Kazazian HH Jr. Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res 2013; 41(15):7401-19; PMID: 23749060; https://doi.org/10.1093/nar/gkt512
- Taylor MS, LaCava J, Mita P, Molloy KR, Huang CR, Li D, Adney EM, Jiang H, Burns KH, Chait BT, et al. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell 2013; 155(5):1034-48; PMID: 24267889; https://doi.org/10.1016/j.cell.2013.10.021
- Moldovan JB, Moran JV. The Zinc-Finger antiviral Protein ZAP Inhibits LINE and Alu retrotransposition. PLoS Genet 2015; 11(5):e1005121; PMID: 25951186; https://doi.org/10.1371/journal.pgen.1005121
- Dai L, Taylor MS, O'Donnell KA, Boeke JD. Poly(A) binding protein C1 is essential for efficient L1 retrotransposition and affects L1 RNP formation. Mol Cell Biol 2012; 32(21):4323-36; PMID: 22907758; https://doi.org/10.1128/MCB.06785-11
- Peddigari S, Li PW, Rabe JL, Martin SL. hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic Acids Res 2013 Jan 7; 41(1):575-85; PMID: 23161687; https://doi.org/10.1093/nar/gks1075
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr. High frequency retrotransposition in cultured mammalian cells. Cell 1996; 87(5):917-27; PMID: 8945518; https://doi.org/10.1016/S0092-8674(00)81998-4
- Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet 2010; 6(10):pii: e1001150; PMID:20949108; https://doi.org/10.1371/journal.pgen.1001150
- Xie Y, Rosser JM, Thompson TL, Boeke JD, An W. Characterization of L1 retrotransposition with high-throughput dual-luciferase assays. Nucleic Acids Res 2011; 39(3):e16; PMID: 21071410; https://doi.org/10.1093/nar/gkq1076
- Heras SR, Macias S, Plass M, Fernandez N, Cano D, Eyras E, Garcia-Perez JL, Cáceres JF. The Microprocessor controls the activity of mammalian retrotransposons. Nat Struct Mol Biol 2013; 20(10):1173-81; PMID: 23995758; https://doi.org/10.1038/nsmb.2658
- Sakurai M, Ueda H, Yano T, Okada S, Terajima H, Mitsuyama T, Toyoda A, Fujiyama A, Kawabata H, Suzuki T. A biochemical landscape of A-to-I RNA editing in the human brain transcriptome. Genome Res. 2014; 24(3):522-34; PMID: 24407955; https://doi.org/10.1101/gr.162537.113
- Goodier JL, Cheung LE, Kazazian HH Jr. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet 2012; 8(10):e1002941; PMID: 23093941; https://doi.org/10.1371/journal.pgen.1002941
- Goodier JL, Pereira GC, Cheung LE, Rose RJ, Kazazian HH Jr. The Broad-Spectrum antiviral Protein ZAP restricts human retrotransposition. PLoS Genet 2015; 11(5):e1005252; PMID: 26001115; https://doi.org/10.1371/journal.pgen.1005252
- Zhang A, Dong B, Doucet AJ, Moldovan JB, Moran JV, Silverman RH. RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res 2014; 42(6):3803-20; PMID: 24371271; https://doi.org/10.1093/nar/gkt1308
- Li P, Du J, Goodier JL, Hou J, Kang J, Kazazian HH Jr, Zhao K, Yu XF. Aicardi-Goutières syndrome protein TREX1 suppresses L1 and maintains genome integrity through exonuclease-independent ORF1p depletion. Nucleic Acids Res 2017; 45(8):4619-31; PMID:28334850; https://doi.org/10.1093/nar/gkx178
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 2003; 35(1):41-48; PMID: 12897783; https://doi.org/10.1038/ng1223
- Zhao K, Du J, Han X, Goodier JL, Li P, Zhou X, Wei W, Evans SL, Li L, Zhang W, et al. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutières syndrome-related SAMHD1. Cell Rep 2013; 4(6):1108-15; PMID: 24035396; https://doi.org/10.1016/j.celrep.2013.08.019
- Goodier JL, Zhang L, Vetter MR, Kazazian HH Jr. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol 2007; 27(18):6469-83; PMID: 17562864; https://doi.org/10.1128/MCB.00332-07
- Goodier JL, Mandal PK, Zhang L, Kazazian HH Jr. Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum Mol Genet 2010; 19(9):1712-25; PMID: 20147320; https://doi.org/10.1093/hmg/ddq048
- Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet 2010; 6(10):pii: e1001150; PMID:20949108; https://doi.org/10.1371/journal.pgen.1001150
- Buchan JR, Parker R. Eukaryotic stress granules: The ins and outs of translation. Mol Cell 2009; 36(6):932-41; PMID: 20064460; https://doi.org/10.1016/j.molcel.2009.11.020
- Weissbach, R, Scadden, AD. Tudor-SN and ADAR1 are components of cytoplasmic stress granules. RNA. 2012; Mar; 18(3):462-71; https://doi.org/10.1261/rna.027656.111.
- Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, Malim MH. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol 2007; 81(5):2165-78; PMID: 17166910; https://doi.org/10.1128/JVI.02287-06
- Hu S, Li J, Xu F, Mei S, Le Duff Y, Yin L, Pang X, Cen S, Jin Q, Liang C, et al. SAMHD1 Inhibits LINE-1 Retrotransposition by promoting stress granule formation. PLoS Genet 2015; 11(7):e1005367; PMID: 26134849; https://doi.org/10.1371/journal.pgen.1005367
- Zheng Y, Lorenzo C, Beal PA. DNA editing in DNA/RNA hybrids by adenosine deaminases that act on RNA. Nucleic Acids Res 2017; pii:gkx050; PMID:28132026; https://doi.org/10.1093/nar/gkx050
- Crow YJ. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Ledbetter N, Mefford HC, Smith RJH, Stephens K, editors. Aicardi-Goutières Syndrome. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2017. 2005 Jun 29 [updated 2016 Nov 22]; PMID:20301648
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 2008; 134(4):587-98; PMID: 18724932; https://doi.org/10.1016/j.cell.2008.06.032
- Volkman HE, Stetson DB. The enemy within: Endogenous retroelements and autoimmune disease. Nat Immunol 2014; 15(5):415-22; PMID: 24747712; https://doi.org/10.1038/ni.2872
- Yu Q, Carbone CJ, Katlinskaya YV, Zheng H, Zheng K, Luo M, Wang PJ, Greenberg RA, Fuchs SY. Type I interferon controls propagation of long interspersed element-1. J Biol Chem 2015; 290(16):10191-9; PMID:25716322; https://doi.org/10.1074/jbc.M114.612374
- Crow MK. Long interspersed nuclear elements (LINE-1): Potential triggers of systemic autoimmune disease. Autoimmunity 2010; 43(1):7-16; PMID:19961365; https://doi.org/10.3109/08916930903374865
- Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, et al. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat Genet 2012; 44(11):1243-48; PMID:23001123; https://doi.org/10.1038/ng.2414

