ABSTRACT
Developmental exposure to endocrine-disrupting chemicals (EDCs), 17β-estradiol-3-benzoate (EB) and bisphenol A (BPA), increases susceptibility to prostate cancer (PCa) in rodent models. Here, we used the methylated-CpG island recovery assay (MIRA)-assisted genomic tiling and CpG island arrays to identify treatment-associated methylome changes in the postnatal day (PND)90 dorsal prostate tissues of Sprague-Dawley rats neonatally (PND1, 3, and 5) treated with 25 µg/pup or 2,500 µg EB/kg body weight (BW) or 0.1 µg BPA/pup or 10 µg BPA/kg BW. We identified 111 EB-associated and 86 BPA-associated genes, with 20 in common, that have significant differentially methylated regions. Pathway analysis revealed cancer as the top common disease pathway. Bisulfite sequencing validated the differential methylation patterns observed by array analysis in 15 identified candidate genes. The methylation status of 7 (Pitx3, Wnt10b, Paqr4, Sox2, Chst14, Tpd52, Creb3l4) of these 15 genes exhibited an inverse correlation with gene expression in tissue samples. Cell-based assays, using 5-aza-cytidine-treated normal (NbE-1) and cancerous (AIT) rat prostate cells, added evidence of DNA methylation-mediated gene expression of 6 genes (exception: Paqr4). Functional connectivity of these genes was linked to embryonic stem cell pluripotency. Furthermore, clustering analyses using the dataset from The Cancer Genome Atlas revealed that expression of this set of 7 genes was associated with recurrence-free survival of PCa patients. In conclusion, our study reveals that gene-specific promoter methylation changes, resulting from early-life EDC exposure in the rat, may serve as predictive epigenetic biomarkers of PCa recurrence, and raises the possibility that such exposure may impact human disease.
KEYWORDS:
- Developmental origin of health and disease (DOHaD)
- early-life reprogramming
- endocrine-disrupting chemicals (EDCs)
- epigenetics
- Ingenuity®
- Pathway Analysis (IPA®)
- methylated-CpG island recovery assay (MIRA)
- NimbleGen rat DNA methylation promoter array
- Sprague Dawley rats
- stem cell pluripotency
- The Cancer Genome Atlas (TCGA)
Abbreviations
| AA | = | African-American |
| Acrbp | = | Acrosin binding protein 21 |
| AIT | = | Rat prostate cancer cell line |
| Akt | = | Protein kinase B |
| 5-aza | = | 5-aza-cytidine |
| BPA | = | Bisphenol A |
| Btbd3 | = | BTB domain containing 3 |
| BW | = | Body weight |
| Ccdc67 | = | Coiled-coil domain containing protein 67 |
| Chad | = | Chondoadherin |
| Chst14 | = | Carbohydrate sulfotransferase 14 |
| Creb3l4 | = | Cyclic AMP responsive element binding protein 3-like 4 |
| Ctrl | = | Control |
| DMR | = | Differentially methylated region |
| Dnase2b | = | Deoxyribonuclease II β |
| Dnmt3a | = | DNA methyltransferase 3a |
| Dnmt3b | = | DNA methyltransferase 3b |
| E2 | = | 17β-estradiol |
| EB | = | 17β-estradiol-3-benzoate |
| EDC | = | Endocrine-disrupting chemical |
| ER | = | Estrogen receptor |
| ERK1/2 | = | Extracellular-signal-regulated kinase 1/2 |
| ERRγ | = | Estrogen related receptor gamma |
| ESR1/ERα | = | Estrogen receptor α |
| ESR2/ERβ | = | Estrogen receptor β |
| GPER1/GPR30 | = | G protein-coupled estrogen receptor 1 |
| Hmgn5 | = | High mobility group nucleosome binding domain 5 |
| Habp2 | = | Hyaluronan binding protein 2 |
| Hpcal1 | = | Hippocalcin-like 1 |
| IPA | = | Ingenuity Pathway Analysis |
| Krt83 | = | Keratin 83 |
| Mbd2 | = | Methyl-CpG binding domain protein 2 |
| Mbd4 | = | Methyl-CpG binding domain protein 4 |
| MIRA | = | Methylated-CpG island recovery assay |
| NbE-1 | = | Rat prostate epithelial cell line |
| Nfia | = | Nuclear factor I/A |
| Osbpl6 | = | Oxysterol binding protein-like 6 |
| Paqr4 | = | Progestin and adipoQ receptor family member 4 |
| PCa | = | Prostate cancer |
| Pde4d | = | Phosphodiesterase type IV variant |
| Phpt1 | = | Phosphohistidine phosphatase 1 |
| Pitx3 | = | Paired-like homeodomain 3 |
| PIN | = | Prostatic intraepithelial neoplasia |
| PND | = | Postnatal day |
| Prkar1a | = | Protein kinase cAMP-dependent type 1 regulatory subunit α |
| Rbpjl | = | Recombination signal binding protein for immunoglobulin kappa J region-like |
| Rnf186 | = | Ring finger protein 186 |
| Rpl19 | = | Ribosomal protein L19 |
| SD | = | Sprague Dawley |
| SEM | = | Standard error of mean |
| Sox2 | = | Sex determining region Y box 2 |
| Tacstd2 | = | Tumor-associated calcium signal transducer 2 |
| Tbx4 | = | T-box 4 |
| TCGA | = | The Cancer Genome Atlas |
| TGFβ | = | Transforming growth factor β |
| Tmem27 | = | Transmembrane protein 27 |
| TNBC | = | Triple negative breast cancer |
| Tpd52 | = | Tumor protein D52 |
| Tymp | = | Thymidine phosphorylase |
| Wnt10b | = | Wingless-type MMTV integration site family, member 10B |
Introduction
Exposure to environmental agents is a risk factor for multiple diseases. Evidence is mounting in support of the causal link between exposure to xenoestrogens and human diseases,Citation1-3 including cancer.Citation4-7 Bisphenol A (BPA) is a ubiquitous environmental xenoestrogen widely used during the production of polycarbonate plastics, epoxy resins, carbonless receipt paper, and hundreds of other manufactured products.Citation1 The fact that urinary BPA is detectable in >95% of the US populationCitation8 indicates that BPA readily leaches into the environment, contaminates our food and water, and enters our bodies. Chronic exposure to BPA in rodentsCitation9 and in humansCitation10 is associated with prostate cancer (PCa) and its pre-lesions. Moreover, BPA is detectable in umbilical cord bloodCitation11-14 and sera of newborns,Citation15 which elevates the concern of early-life BPA exposure and development of later-life disease. As an endocrine disrupting chemical (EDC), BPA binds to estrogen receptors (ERs) ESR1, ESR2, and GPER1,Citation16 which are expressed in the rodent and human prostate epithelial cells.Citation17,18 While affinity for nuclear ERs is low compared to estradiol-17β, BPA has equivalent activational capacities for membrane ERsCitation16,19 and rapid actions of low-dose BPA have been documented in prostate cells.Citation19 Finally, BPA can also signal through non-ER pathways.Citation20,21
Like BPA, 17β-estradiol-3-benzoate (EB) is a ubiquitous environmental agent. It is an estradiol analog commonly used in livestock for inducing weight gain and synchronizing estrus cycles in heifers and cattle.Citation22 Since EB binds ERs with high affinity,Citation23 unintended exposures may trigger estrogenic responses that compromise health. In rodents, neonatal EB exposure (30–125 ng/day in mouse and 125–500 ng/day in rats) has been shown to induce the production of morphologically abnormal sperm during adulthood.Citation24 Similarly, rats neonatally exposed to 25 µg EB developed prostates with a greater risk of malignant changes with aging, including severe prostatic intraepithelial neoplasia (PIN), a precancerous condition.Citation25-27 Since production of morphologically abnormal spermCitation24 and induction of increased susceptibility to hormonal carcinogenesisCitation25 are similarly observed following neonatal BPA exposure, these findings strongly implicate early-life as a susceptible window for environmental xenoestrogens to predispose later-life disease risk and EB can serve as a positive estrogen control in studies of weaker estrogenic agents such as BPA. While emerging evidence indicates that epigenetic modifications are involved,Citation25,28-33 the molecular underpinnings of this process remain to be fully clarified.
To initially interrogate whether neonatal exposure to EB and BPA reprogrammed the prostate epigenome, we exposed rats to an environmentally relevant dose of BPA [10 μg/kg body weight (BW)] or EB (2,500 μg/kg BW) on postnatal days 1, 3, and 5. We observed that this transient developmental exposure increased the dorsal and lateral prostate lobe susceptibility to adult-onset E2-induced carcinogenesis, markedly augmenting the incidence and severity of lesions as compared to oil-treated controls.Citation25,32,34 Using methylation-sensitive restriction fingerprinting to identify altered DNA methylation marks, we found that neonatal EB and BPA exposures were associated with the dysregulation of phosphodiesterase type IV variant 4 (Pde4d4),Citation25 hippocalcin-like 1 (Hpcal1; also known as visinin-like protein-3, Vilip-3), and high mobility group nucleosome binding domain 5 (Hmgn5; formerly known as nucleosome binding protein 1, Nsbp1)Citation32 through aberrant promoter methylation detected on postnatal day (PND)10, 90, and 200. Together, these findings provided the first evidence that early–life environmental exposure to EDC reprogrammed the prostate epigenome and identified a developmental basis of PCa risk with aging. Moreover, the expression of DNA methylation transferase3a and 3b (Dnmt3a and Dnmt3b) and methyl-CpG binding domain protein2 and 4 (Mbd2 and Mbd4) was upregulated in PND10 and 90 prostate tissues,Citation32 providing a mechanistic basis for reprogramming of DNA methylation marks upon early-life EDC exposure. Thus, we propose that differential methylation of genes that persist in the PND90 prostate primes the tissue for heightened sensitivity to a secondary exposure to rising estradiol later in life, as occurs in aging males.Citation35 This is particularly relevant since elevated estrogens have been associated with increased PCa risk in menCitation6 and are sufficient to transform the human prostate epithelium.Citation17
While these findings had a major impact on the field of EDC research and developmental basis of carcinogenesis,Citation36 a genome-wide search for additional DNA methylation targets of neonatal exposure to xenoestrogens is warranted for an unbiased discovery of other epigenetic marks that may underlie increased PCa risk with aging. To accomplish this goal, we herein used the methylated-CpG island recovery assay (MIRA)-assisted genomic tiling and CpG island array analysis and identified distinct and common EB-/BPA-associated genes in PND90 prostate tissues from rats with neonatal exposure to these xenoestrogens. Among 25 epigenetically regulated candidate genes, the promoter methylation status of 7 genes (Pitx3, Wnt10b, Paqr4, Sox2, Chst14, Tpd52, Creb3l4) was inversely correlated to gene expression. These validated genes have functional connectivity associated with stem cell pluripotency. Of clinical relevance, expression of these genes was found to be associated with recurrence-free survival of 497 patients in The Cancer Genome Atlas (TCGA) PCa cohort, suggesting that they may have utilities for predicting disease progression and patient stratification based on disease aggressiveness.
Results
Methylation array revealed differential promoter methylation of genes associated with neonatal EB or BPA exposure in the PND90 prostate
We performed genome-wide methylation analysis, using MIRA-assisted genomic tiling and CpG island array, in dorsal prostate tissues from PND90 rats neonatally exposed to EB or BPA (). Using a 750 bp sliding window approach and a selection criteria of P < 10−5 and mBar >0.6 or <−0.25 (), we identified a total of 177 differentially methylated regions (DMRs), which were randomly distributed among chromosomes (Supplemental Figure S1). Since this array was designed to target CpG sites only at the gene promoter region, the genes described hereafter refer to those identified with DMRs at the 5′-promoter region. Of the 177 genes with identified DMRs, 111 genes were EB-associated, 86 genes were BPA-associated, and 20 genes were common between the 2 groups (; Supplemental Table S1).
Figure 1. Schematic diagram of the experimental and analytical procedures. Sprague-Dawley (SD) rats were neonatally [postnatal day (PND) 1, 3, 5] treated with 17β-estradiol-3-benzoate (EB) at 25 μg/pup or 2,500 µg/kg body weight (BW), bisphenol A (BPA) at 0.1 µg/pup or 10 µg/kg BW, or corn oil as a control (Ctrl). DNA extracted from PND90 dorsal prostate tissue was subjected to promoter methylation array analysis, and genes with differentially methylated regions and inverse expression correlation were identified and validated.
![Figure 1. Schematic diagram of the experimental and analytical procedures. Sprague-Dawley (SD) rats were neonatally [postnatal day (PND) 1, 3, 5] treated with 17β-estradiol-3-benzoate (EB) at 25 μg/pup or 2,500 µg/kg body weight (BW), bisphenol A (BPA) at 0.1 µg/pup or 10 µg/kg BW, or corn oil as a control (Ctrl). DNA extracted from PND90 dorsal prostate tissue was subjected to promoter methylation array analysis, and genes with differentially methylated regions and inverse expression correlation were identified and validated.](/cms/asset/3aaeca16-9043-45ff-97a8-1f4b2d708db0/kepi_a_1208891_f0001_c.gif)
Biological significance of differentially methylated regions associated with neonatal EB and BPA exposure in PND90 prostate
To identify biological processes related to the identified exposure-associated DMRs, we performed functional network analysis using Ingenuity Pathway Analysis (IPA). As shown in , the top 3 networks related to the EB-associated genes were identified as: 1) “tissue morphology, embryonic development, organ development;” 2) “cell-to-cell signaling and interaction, cell-mediated immune response, cellular growth and proliferation;” and 3) “post-translational modification, cellular assembly and organization, cellular function and maintenance.” Upon merging the molecular interactions of these top 3 networks, the EB-associated genes were found to converge at protein kinase B (AKT) and extracellular-signal-regulated kinase (ERK)1/2. While the EB-associated genes had implications in the development and function of several physiological systems, including embryonic, tissue, organ, and reproductive (), the top related disease was “cancer,” which included PCa.
Table 1. Top networks and bio-functions of the genes associated with neonatal 17β-estradiol-3-benzoate (EB) or bisphenol A (BPA) exposure in postnatal day (PND)90 prostate.
Similarly, the top 3 networks related to the 86 BPA-associated genes were identified as: 1) “cell-to-cell signaling and interaction, cell-mediated immune response, cellular growth and proliferation;” 2) “nucleic acid metabolism, small molecule biochemistry, molecular transport;” and 3) “cellular assembly and organization, cellular function and maintenance, cellular compromise” (). The molecular interactions of these networks converged at transforming growth factor β (TGFβ), and the 86 genes had implications in digestive and cardiovascular system development and function, organ and tissue morphology, and organismal development (). Like the EB-associated genes, BPA-associated genes were also highly associated with “cancer,” including PCa.
Interestingly, the top 3 networks related to the 20 associated genes common of both EB- and BPA-exposure were: 1) “cancer, organismal injury and abnormalities, renal and urological disease;” 2) “cell-to-cell signaling and interaction, nervous system development and function, cardiovascular disease;” and 3) “cancer, connective tissue disorders, dermatological diseases and conditions” (Supplemental Table S2C). While these 3 networks converged at tumor protein 53 (TP53), overall, the 20 genes were significantly related to digestive, hepatic, and nervous system development and function, and embryonic and organismal development (Supplemental Table S2). They also had implications in “connective tissue disorders,” “developmental disorder,” “hereditary disease,” “inflammatory disease,” and “metabolic disease” (Supplemental Table S2).
Selection of top candidate genes for methylation validation by bisulfite sequencing
To tighten the selection of differentially methylated candidate genes, we initially sorted the 177 identified genes based on P < 10−10 and mBar (EB/BPA – Control) ≥1 .5 or ≤−0.5 (Selection criteria A, ). Under this stringent selection criteria, 20 candidate genes were identified (Supplemental Table S3), of which the top 9 most differentially methylated genes, based on differences in mBar values (), across exposure groups (Btbd3, Chst14, Creb3l4, Paqr4, Phpt1, Rbpjl, Sox2, Tacstd2, and Tpd52) were selected for validation. As shown in , primers for bisulfite sequencing validation for each gene were designed to amplify the region with the greatest methylation changes between the EB/BPA-treated groups and the control group. In most cases, the interrogated regions were closely aligned to the predicted CpG island based on MethPrimer program.Citation37
Figure 2. Representative results from genome-wide methylation study. A) Predicted CpG islands (light blue shaded areas) in the promoter region of differentially methylated genes (Pitx3, Wnt10b, Paqr4, Sox2, Chst14, Tpd52, and Creb3l4) identified in this study. TSS stands for transcriptional start site whereas ATG stands for translational start codon. Individual CpG sites are represented by red vertical lines. Dark blue horizontal line marks the region selected for BS-sequencing; B) Position of the BS-sequenced region (blue line) relative to the NimbleGen probes covered regions (dirty yellow lines) of each gene; C) Significant methylation level of each gene in vehicle- (control), EB- and BPA-treated groups, which were measured by NimbleGen array probes (mBar values). The height of the mBar represents the probe intensity; red and green bars represent positive and negative methylation value, respectively, relative to their respective input control.
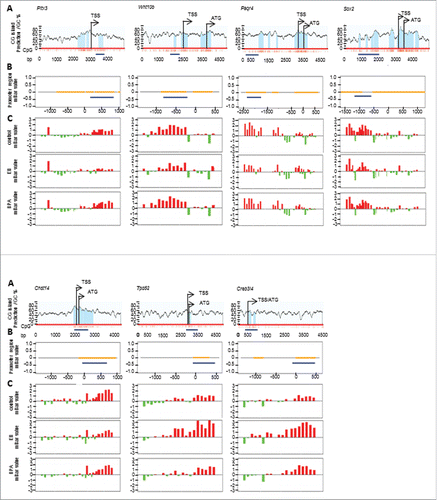
Promoter methylation status was validated by performing bisulfite sequencing analysis on the same PND90 prostate tissue samples used in the methylation array (; Supplemental Table S3). In accordance with the methylation patterns observed in the array, the DMRs of 7 out of 9 genes were confirmed. The DMRs of Chst14, Paqr4, Phpt1, Rbpjl, and Sox2 were hypomethylated, while those of Creb3l4 and Tpd52 were hypermethylated in the EB/BPA-exposure groups when compared to controls ().
Table 2. Validation of promoter methylation status and gene expression in postnatal day (PND)90 prostate.
To include more genes in the analysis, we also sorted the 177 exposure-associated genes using wider selection parameters of P < 10−5 and mBar (EB/BPA – Control) ≥1.2 or ≤−0.5 (Selection criteria B, ). Using selection criteria B, 91 candidate genes were identified (Supplemental Table S4). Bisulfite sequencing analysis was performed on the top 16 most differentially methylated genes across exposure groups (Acrbp, Ccdc67, Chad, Dnase2b, Habp2, Krt83, Nfia, Osbp16, Pitx3, Prkar1a, Rnf186, Tbx4, Tmem27, Tymp, Uox, and Wnt10b) (; Supplemental Table S4). Promoter methylation validation of the 16 genes, using PND90 dorsal prostate tissues, confirmed promoter hypomethylation of Pitx3 and Wnt10b, and promoter hypermethylation of Acrbp, Chad, Osbp16, Rnf186, Tmem27, and Tymp ().
Overall, using 2 selection criteria, there were 25 top differentially methylated candidate genes identified, of which the promoter methylation status of 15 were validated (Chst14, Creb3l4, Paqr4, Phpt1, Rbpjl, Sox2, and Tpd52 from selection criteria A; and Acrbp, Chad, Osbp16, Pitx3, Rnf186, Tmem27, Tymp, and Wnt10b from selection criteria B) ().
Validation of promoter methylation status and gene expression correlation
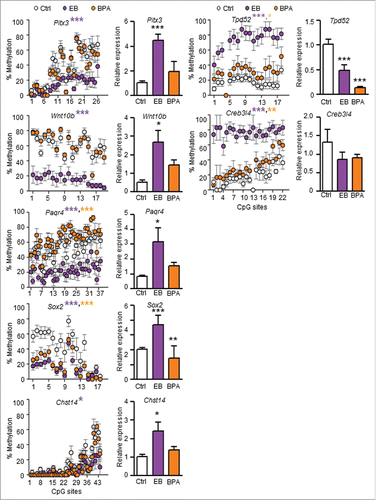
To elucidate the biological relevance of the DMRs within these gene promoters, we next performed gene expression analyses on the PND90 prostate tissues. As shown in , expression of Pitx3, Wnt10b, Paqr4, Sox2, and Chst14 was significantly upregulated in the EB-exposure group when compared to the control. Of these genes, only Sox2 was significantly upregulated in the BPA-exposure vs. control group. In contrast, expression of Tpd52 was significantly downregulated in both exposure groups, and Creb3l4 showed a trend of downregulation in exposure groups, when compared to the control group (). Since gene expression of Acrbp, Chad, Osbp16, Rnf186, Tmem27, and Tymp in the prostate tissues of the exposure groups did not associate with their validated promoter methylation status, they were not studied further (; data not shown).
Figure 3. Effect of neonatal exposure to EB or BPA on promoter methylation and gene expression in PND90 dorsal prostate. Promoter methylation status (left panel: Scatter Plot) and expression (right panel: Bar Graph) of candidate genes, in PND90 prostate tissues from SD rats treated with either corn oil (Ctrl; white), EB (green), or BPA (red), were analyzed using bisulfite sequencing and qPCR, respectively. Each circle in the scatter plot represents mean ±SEM of methylation percentage (averaged from 6 individual samples/animals) at a single CpG site in the gene promoter region. The % methylation of each CpG site in each sample was determined from bisulfite sequencing data derived from 8–12 clones. Gene expression data were expressed as mean ± SEM from 3 individual samples. Statistical significance was determined by one-way ANOVA and Tukey test when compared to Ctrl. *P < 0.05, **P < 0.01, and ***P < 0.001.
To investigate the collective biological relevance of the 7 candidate genes (Pitx3, Wnt10b, Paqr4, Sox2, Chst14, Tpd52, and Creb3l4), we performed functional network analysis using IPA. We found that the candidate genes were associated with 2 networks: 1) “embryonic development, organismal development, gene expression” and 2) “cell cycle, embryonic development, gene expression” (Supplemental Table S5), which converged at Sox2 (). While these 7 candidate genes had implications in embryonic, organ, and organismal development, as well as auditory, vestibular, and nervous system development and function, “cancer” was determined to be the top-related disease associated with the differentially methylated genes with inverse expression correlation (Supplemental Table S5). Complementary reported bio-functions are summarized in .
Figure 4. Signaling pathways associated with differentially methylated candidate genes were predicted by Ingenuity Pathway Analysis™. Genes with promoter hypermethylation are shown in red and genes with promoter hypomethylation are shown in green, with color intensity signifying the magnitude of differential methylation. Gray arrows indicate predicted association pathways; purple arrows indicate reported directional pathways.
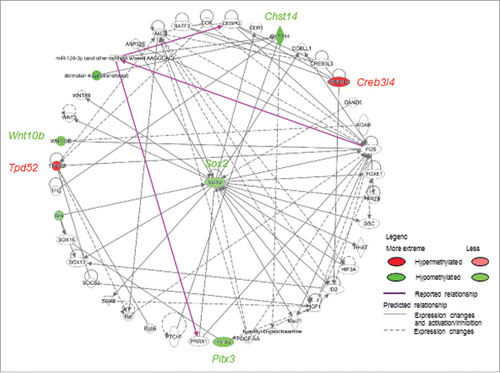
Table 3. Reported bio-function(s) and PCa associations of the seven identified candidate genes with DMRs inversely correlated with gene expression as a result of neonatal 17β-estradiol-3-benzoate/bisphenol A (EB/BPA) exposure.
Effect of 5-aza-cytidine on the expression of differential methylation candidate genes in rat prostate cells
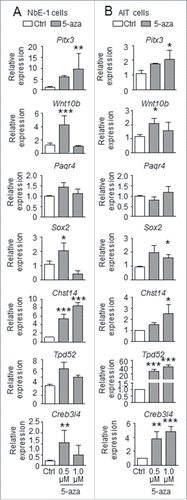
To determine whether the expression of the 7 identified candidate genes (Pitx3, Wnt10b, Paqr4, Sox2, Chst14, Tpd5, and Creb3l4) were directly regulated by DNA methylation, we compared gene expression in a rat prostate epithelial cell line NbE-1 Citation38 and the rat PCa cell line AITCitation39, either with or without a 4-day treatment of the DNA methylation inhibitor, 5-aza-cytidine (5-aza) at doses of 0.5 µM and 1 µM. When compared to the untreated control, 5-aza treatment significantly upregulated Pitx3, Wnt10b, Chst14, and Creb3l4 in NbE-1 cells, and Pitx3, Wnt10b, Sox2, Chst14, Tpd52, and Creb3l4 in AIT cells (). Although the same treatment increased the expression of Sox2 and Tpd52 in NbE-1 cells, the change was not significant. Overall, the 5-aza treatment had no effect on the expression of Paqr4 in both NbE-1 and AIT cells.
Figure 5. Effect of 5-aza-cytidine treatment on gene expression in NbE-1 and AIT cells. Gene expression was analyzed by qPCR in rat (A) normal prostate epithelial NbE-1 cells and (B) prostate cancer AIT cells treated with DMSO (Ctrl), or 0.5 µM or 1 µM 5-aza-2-deoxycytidine (5-aza), a DNA methylation inhibitor, for 8 d. Data (mean ± SEM ) is normalized to corresponding Rpl19 levels, and is expressed as fold change vs. Ctrl. *P < 0.05, and **P < 0.001 by one-way ANOVA and Tukey test when compared to Ctrl.
Association of the identified candidate genes with the recurrence of PCa
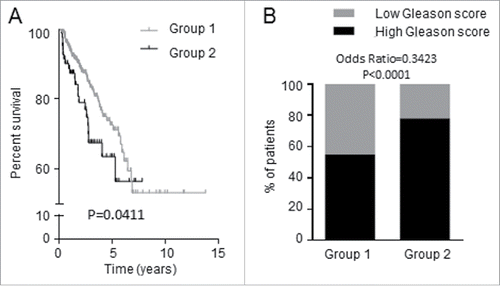
To gain clinical significance, RNAseq data from 497 PCa specimens and their associated clinical data (Biotab) were retrieved from the TCGA database. Extensive data transformation and biostatistical analyses revealed that the expression of these 7 genes in human PCa samples correlated with recurrence-free survival, i.e., the cohort of patients was segregated into those with longer (Group 1) and shorter (Group 2) duration to cancer re-appearance after initial treatment. For the initial 5-year survival after diagnosis, Group 2 patients had a shorter time to recurrence (poorer recurrence-free survival) when compared with Group 1 (, Hazard ratio = 1.786, log rank test P = 0.0411), suggesting Group 2 had significantly more aggressive disease. Consistent with this finding, we found that Group 2 had significantly more cases with higher tumor Gleason score (≥7) when compared with Group 1 (Odds ratio = 0.3423; P < 0.0001) (). It is well established that men with higher grade tumors have higher risk of dying from PCa.Citation40
Figure 6. Expression of the 7 candidate genes was associated with shorter recurrence-free survival of PCa patients. TCGA data consisting of 497 PCa patients were dichotomized into Groups 1 and 2 by K-means clustering analysis based on the 7 candidate genes. (A) Group 1 patients have longer time to recurrence than Group 2 patients; the two groups differ in recurrence-free survival. (B) Proportion of PCa patients having tumors with high (≥7) and low (<7) Gleason score.
Discussion
In the present study, we conducted a comprehensive genome-wide search for DNA methylation targets of developmental exposure to EB and BPA, with a view to identifying candidate epigenetic biomarkers for predicting PCa risk. We reported the genome-wide promoter methylation changes in PND90 dorsal prostate tissues related to neonatal exposure to EB or BPA. Overall, we identified 111 EB-associated and 86 BPA-associated DMRs, with 20 in common between the 2 groups. Using two selection criteria, 25 candidate genes were selected, of which 15 were validated to have treatment-associated promoter methylation changes using bisulfite sequencing analysis, and 7 identified to exhibit an inverse correlation between promoter methylation status and gene expression. EB treatment upregulated the expression of Pitx3, Wnt10b, Paqr4, Sox2, and Chst14 through promoter hypomethylation, and downregulated Tpd52, and Creb3l4 through promoter hypermethylation. Similarly, BPA treatment upregulated Sox2 and Chst14 through promoter hypomethylation, and downregulated Tpd52 through promoter hypermethylation, but had no significant effects on the other genes. Of the 7 candidate genes identified in the rat, WNT10b, SOX2, TPD52, and CREB3L4 were upregulated in PCa (), and PAQR4, WNT10b, PITX3, and TPD52 were differentially methylated between PCa and adjacent normal prostate tissues.Citation69 Clustering analysis, based on the expression of these 7 genes in PCa tissues of 497 PCa patients from the TCGA data sets, further revealed an association with shorter recurrence-free survival. Taken together, DNA methylation-mediated dysregulation of this unique signature of 7 genes established a causal link between early-life EDC exposure and PCa risk, and raises the possibility that such exposure may impact the human disease.
Neonatal development is a susceptible window for the DNA methylation reprogramming effects of EDCs. Since the prostate is estrogen responsive, it is highly vulnerable to insult by estrogenic agents during development.Citation27,34,41 Using a neonatal EB/BPA exposure rat model, we previously reported that treatment-associated dysregulation of Pde4d4, Hmgn5, and Hpcal1 through promoter methylation changes, persisted in the prostate throughout life.Citation25,32 While we observed an increase in the incidence and severity of PIN in PND200 rats upon E2-induced carcinogenesis,Citation25,32 the current study aimed to interrogate the underlying mechanism for the early-life EDC exposure-associated increase in PCa risk, by performing genome-wide promoter methylation analysis on PND90 prostate tissue. The 111 EB- and 86 BPA-associated genes with identified DMRs were mostly related to cell-to-cell signaling and interaction, cell-mediated immune response, and cellular growth and proliferation, with cancer as the major disease outcome. Specifically, most of these treatment-associated genes were related to 2 oncogenic rapid signaling molecules AKT and ERK1/2. AKT is upregulated in prostate tumors when compared to benign prostatic tissuesCitation42,43 and is associated with a stage-dependent increase in tumor cell proliferation.Citation44 Its activation has also been shown to suppress androgen/androgen-receptor induced apoptosis in PC3 cells.Citation45 Unlike AKT, expression of ERK1/2 is similar between PCa and normal tissues, yet its activation is associated with increased cell proliferation in PCa tissues.Citation46 Moreover, the concomitant activation of Akt and Erk1/2 promotes PCa cell growth and tumorigenicity in a rodent PCa model.Citation47 Taken together, these reports suggest a possible role for the identified treatment-associated genes in the malignant transformation of rat prostate epithelial cells.
Interestingly, functional connectivity of the 7 candidate genes was linked to embryonic stem cell pluripotency, further evidenced by convergence at Sox2. SOX2 is a stem cell marker upregulated in PCa.Citation48-50 Overexpression of SOX2 in human DU145 PCa cells promotes tumorigenesis,Citation48 increases cell proliferation and migration, and reduces apoptosis.Citation51,52 Of the other identified genes, TPD52, an oncogene, has been reported to be upregulated in high-grade PINCitation33,53 and PCa.Citation52-54 CREB3L4Citation55,56 and WNT10BCitation57 have also been found to be upregulated in prostate tumors. On the contrary, the expression of PITX3 and CHST14 has yet to be studied in PCa tissues. While only 4 of the 7 genes have been functionally characterized in PCa cells and tissues, all genes, except PAQR4, are related to stem cell proliferation, cancer cell proliferation migration, and/or tumorigenesis (reported bio-functions are summarized in ).
We have previously shown that prostate stem/progenitor cells isolated from the human adult prostate are more proliferative and able to retain their “stemness” properties when treated with E2 or BPA.Citation17,19 Suppression of a class of non-coding RNAs, the small nucleolar RNA C/D box,Citation30 associated with distinct histone modificationsCitation19,30 has been postulated as a mechanism causing a slow-down in biosynthesis and stem cell differentiation, thus extending the proliferation phase of these cells. In concordance, our current study showed that neonatal exposure to xenoestrogens led to the dysregulation of a panel of stem cell function-related genes through DNA methylation in the adult prostate, lending further credence to this hypothesis. In this regard, neonatal exposure to xenoestrogens may allow for an extended proliferation phase and increase the number of stem/progenitor cells in the prostate. As such, these cells are known targets for carcinogen-induced malignant transformation in adult-life, leading to increased cancer susceptibility.Citation58
In addition to their reported involvement in PCa, of clinical relevance, we report here that the expression of the novel 7-gene set is associated with shorter recurrence-free survival of PCa patients in the TCGA cohort. These patients were also mostly diagnosed with prostate tumors of high Gleason score (≥7). Of interest, we also found that 4 out of these 7 genes were differentially methylated between PCa tissues and their adjacent normal tissues in the cohort reported by Kim et al.Citation59 Thus, this gene signature may have diagnostic/prognostic value by serving as biomarkers for predicting disease initiation and progression.
Although neonatal exposure to either EB or BPA increased PCa risk in the rodent model, their DNA methylation mediated effects on the prostate are different. There were more EB-associated than BPA-associated genes identified from our promoter methylation array. Genes with methylation changes validated were also mostly EB-associated. Moreover, the EB-associated genes linked to AKT and ERK1/2 as the major signaling molecules, whereas the BPA-associated genes linked to TGFβ, an oncogenic molecule upregulated in PCa.Citation60 This indicates that they activate different signaling pathways to increase PCa risk. Furthermore, EB-treatment dysregulated Pitx3, Wnt10b, Paqr4, and Creb3l4 in PND90 prostate tissues through promoter methylation changes, but BPA exposure had no significant effects. This suggests that EB had a stronger effect on the prostate than BPA, which could in part be related to a markedly higher dose of EB utilized (2500 µg/kg BW) when compared to the lower environmentally relevant dose of BPA (10 µg/kg BW). Alternatively, it has been reported that the action of BPA can be independent of the classical estrogen receptors (ERα and ERβ) and mediated via estrogen-related receptor γ (ERRγ) and G protein-coupled receptor 30 (GPER1), to name a few.Citation61,62 However, this does not explain why the expression of Acrbp, Chad, Osbpl6, Phpt1, and Tmem27 did not correlate to their confirmed promoter methylation status, nor why the expression of Paqr4 was unaffected by 5-aza treatment in NbE-1 and AIT cells. In human prostaspheres, E2/BPA-mediated small nucleolar RNA box C/D suppression was shown to be associated with the occupancy of histone marks the trimethylated histone 3 at lysine 9 and lysine 27, and the loss of trimethylated histone 3 at lysine 4, rather than DNA methylation.Citation30 Thus, EB and BPA may dynamically alter the PND90 prostate transcriptome through other epigenetic mechanisms not yet explored.
In summary, we have identified a set of 7 genes (Pitx3, Wnt10b, Paqr4, Sox2, Chst14, and Tpd52, and Creb3l4) with differential methylation at the promoter region, in PND90 dorsal prostate tissue of rats neonatally exposed to EDCs. The methylation pattern of these 7 genes was inversely correlated to gene expression. Cell-based studies using 5-aza-cytidine-treated normal (NbE-1) and cancerous (AIT) prostate epithelial cell lines further confirmed that gene expression was regulated by DNA methylation, in 6 of the 7 genes. To provide clinical relevance, 4 of the 7 genes (PITX3, WNT10B, PAQR4, and TPD52) have been reported to be differentially methylated between PCa and normal adjacent tissues. Moreover, expression of the 7 genes is associated with the recurrence-free survival of PCa patients. Taken together, we conclude that DNA methylation changes in our novel 7-gene signature may be an epigenetic signature associated with increased cancer susceptibility in the adult gland due to early-life exposure.
Materials and methods
Animal housing and treatments
All animal treatments in this study were in accordance with the accepted standards of humane animal care and approved by the Animal Use Committee at the University of Illinois. Pregnant Sprague-Dawley (SD) rats used on postnatal day (PND) 90 were from Zivic-Miller Laboratories (Pittsburgh, PA) and shipped on gestational day 12. Animal husbandry and conditions were controlled to avoid inadvertent BPA and phytoestrogen exposures, as reported previously.Citation21,63
A schematic diagram of the experimental and analytical procedures is outlined in . Pregnant dams were monitored and the day of birth was designated as PND0. Anogenital distance was used to segregate male and female pups. Male pups were randomly assigned to one of 3 treatment groups, n = 30/group: (a) controls given tocopherol-stripped corn oil vehicle alone (Ctrl), (b) 25 µg EB/pup or 2,500 µg EB/kg BW (Sigma-Aldrich Chemical Co., E8515), or (c) 0.1 µg BPA/pup or 10 µg BPA/kg BW (Sigma-Aldrich Chemical Co., 239658). To avoid litter effects, pups from each litter were equally divided among treatment groups and toe clipped for permanent identification. Pups were subcutaneously injected in the nape of the neck with the respective steroids on PND1, 3 and 5, weaned on PND21, and siblings were housed until PND90. The dose and route of EB and BPA was based on our published studyCitation25,64 and utilized to maintain continuity of results.Citation27 We had previously characterized the BPA pharmacokinetics in PND3 neonates following a single subcutaneous injection of 10 µg/kg BW of the endocrine disruptor.Citation34 The mean sera unconjugated BPA level reached a Cmax of 1.77 ng/ml in 0.5 h but rapidly declined to 0.7 ng/ml and 0.54 ng/ml at 1 h and 2 h, respectively, post-injection. These findings suggest a rather rapid rate of bio-degradation of BPA in PND3 neonates, making it unlikely that BPA will reach higher levels upon multiple injections (PND1, 3, 5). However, since we did not have actual measurements of circulating BPA in the neonates during this period, this conclusion may simply be a conjecture. Yet, it should be noted that these levels and those reported before in rodent neonatesCitation34,65 are well within the range reported for human developmental exposures.Citation12,15,34
Genomic DNA extraction from dorsal prostate
Genomic DNA was extracted from the rat dorsal prostate using DNeasy Blood & Tissue kit (Qiagen, Valencia, CA) and in the presence of RNase A (Qiagen 19101) according to the manufacturer's protocol. All reagents were supplied in the kit. In brief, the tissue was lyzed in Buffer ATL with proteinase K at 56°C until the tissue was completely lyzed. The lysate was incubated with 100 mg/ml RNase A and mixed thoroughly with equal portions of Buffer AL and 100% ethanol. The mixture was loaded on to a DNeasy Mini spin column, and washed with Buffer AW1 and Buffer AW2 consecutively by centrifugation. Genomic DNA was eluted with DEPC water diluted Buffer AE.
Methylated CpG island Recovery Assay (MIRA)-assisted Methylation array analysis
A total of 5, 4, and 5 PND90 prostate genomic DNA samples from EB, BPA, and Ctrl group, respectively, were used to perform the MIRA-assisted genome-wide methylation analysis. In brief, 4.5 µg genomic DNA was sonicated by Bioruptor (Diagenode, Belgium) to obtain 300–500 bp fragments. The methylated DNA fraction was enriched using MethylCollector Ultra kit (ActiveMotif, 55005) via MIRA-assisted methylated DNA enrichment with proper enrichment controls. It should be noted that the MIRA-, also known as MBD2-, assisted procedure enriches CpG-dense sequences that are likely in CpG islands.Citation66 The methylation-enriched DNA and its corresponding Input were then amplified using GenomePlex WGA kit (Sigma-Aldrich, WGA2), labeled with Cy3/Cy5 dyes, and co-hybridized in Rat ChIP 385K Promoter 2 array set (Roche NimbleGen, Madison, WI) according to the manufacturer's protocol. The arrays have ∼780,000 probes with probe sizes ranging from 50–70 mers in length and median probe spacing of 105 bp, covering ∼16,000 regions primarily in promoter regions of ∼23,000 transcripts from known genes. These regions principally contain CpG-dense DNA sequences. The probes are typically placed at 4,500 bp upstream and 1,125 bp downstream from the transcriptional start site (TSS). The estimated coverage approximates 97Mb and 3.7% coverage of the rat genome.Citation67 Thus, the design of the experiments was to focus on CpG rich regions within or near known or predicted gene promoters. This design has the advantage of increasing the hit rate and reducing noise, but does have the deficit of biasing toward gene promoters and having low coverage of the overall rat genome and DMRs not assisted with gene promoters.
Methylation array data analysis
Data analysis was performed using R and Bioconductor packages.Citation68 The log-ratios of Cy5 to Cy3 (M values) were first normalized using GC-loess normalization. The average M value was then calculated for each probe across all samples in the same group. The significantly enriched regions were identified using sliding window analysis for each group separately. In this analysis, a window of 750 bps around each probe was considered at a time and RandomSet statisticCitation69 was calculated on the average M values of all probes in the window to estimate the enrichment of the probes in the window when compared to the background. Since there is no enrichment of immunoprecipitated DNA among the probes in the window, the RandomSet statistic, which is defined as the average of the average M values in the window, is approximately distributed as the normal random variable. The P-value of each window was obtained based on the RandomSet statistic and adjusted by the false discovery rate. Windows that were significantly enriched (P < 10−5) in any of the groups (EB, BPA, and Ctrl) were mapped to genes and were further filtered, where differentially methylated genes were selected based on the difference of mBar values of treatment group (EB/BPA) when compared to Ctrl and at different cutoff levels of P-value (). Using this approach, a methylation bar plot was generated for each promoter region in the 3 groups (). In a given promoter region, the difference in height of the bars (mBar) between treated and Ctrl groups represents the difference in probe intensity, i.e., the difference in promoter methylation status. When compared with the control, a positive and negative mBar difference in the treated group indicates promoter hyper- and hypo-methylation, respectively. Primers were then designed based on these differentially methylated regions for validation using bisulfite sequencing analysis.
Bisulfite PCR sequencing analysis
Based on the methylation array bar plot, significant differential methylated regions flanking the gene transcription start sites were selected for methylation validation using bisulfite sequencing. Primers used for bisulfite sequencing were designed with MethPrimer.Citation37 (Supplemental Table S6) Genomic DNA was bisulfite modified using an EZ DNA Methylation kit (Zymo Research, D5001). In brief, 500 ng genomic DNA diluted with M-Dilution buffer was incubated at 37°C for 15 min, mixed with CT reagent and incubated for 16 hours at 50°C. The reaction was then mixed with Binding buffer, loaded to a spin column, washed, incubated with desulphonation buffer at room temperature for 20 min, and eluted with 40 µl elution buffer. Bisulfite PCR was performed using 2 µl bisulfite modified DNA and Platinum Taq DNA Polymerase (Invitrogen, 10966026) in a 25 µl reaction according to the manufacturer's protocol. Following 40 cycles of PCR amplification with the annealing temperature at 55°C, the amplicons were gel-purified and TA-cloned into pGEMT-easy vector (Promega, A1360). Plasmids from a single E. coli colony were amplified using TempliPhi DNA amplification kits (GE Healthcare, 25640010) and sequenced (Macrogen USA, Rockville, MD). The methylation status of each CpG site was analyzed using BiQ Analyzer.Citation70
To validate the promoter methylation status of genes identified from the PND90 prostate microarray analysis, the same amount of DNA from each animal of the same treatment group was mixed for bisulfite sequencing with ∼12 clones per group. To further compare methylation pattern of promoter DMR of the 7 selected gene candidates, bisulfite sequencing analysis on 6 samples each from individual animal was performed with ∼8–12 clones selected per sample.
Treatment of NbE-1 and AIT cells
Rat prostatic epithelial cell lines, the immortalized normal prostatic epithelial cell line NbE-1Citation38 and the tumorigenic cell line AITCitation39 were maintained in DMEM/F12 medium (Invitrogen, 11330057) supplemented with 5% fetal bovine serum (HyClone, 35-010-CV), 1X insulin-transferrin-selenium (Invitrogen, 41400045), 1 mM sodium pyruvate (Invitrogen, 11360070), and 100 mM minimum nonessential amino acids (Invitrogen, 11140076). The cells were seeded and treated with 0.5 µM or 1.0 µM 5-aza-2-deoxycytidine (5-aza-dC; Tocris, 2624), a DNA methylation inhibitor, every 2 d for 8 d as previously described.Citation25,32 Treatment with 0.1% dimethylsulfoxide (DMSO; Sigma D4540) was used as control (Ctrl) in each experimental set. Cell lysates were stored in TRIzol reagent for RNA extraction.
RNA extraction and real-time RT-qPCR
Dorsal prostate tissue was homogenized in TRIzol reagent (Invitrogen, 15596) using Precellys 24 homogenizer (Bertin Technologies, France) and extracted according to the manufacturer's protocol. One microgram total RNA was reversed transcribed using SuperScript III Reverse Transcriptase (Invitrogen, 18080085) in a 20 µl reaction. Primers for real-time RT-qPCR were designed using Primer-BLAST program (Supplemental Table S7). qPCR analysis was performed in a 7500 Fast Real-Time System (ABI, Foster City, CA) in triplicate in a 20 µl reaction. Target gene expression was normalized against the individual endogenous Rpl19Citation34 and relative gene expression was calculated using the 2−ΔΔCt method.Citation30 Similarly, total RNA from treated cells was extracted using TRIzol reagent (Invitrogen, 15596-018) according to the manufacturer's protocol and reversed transcribed for qPCR analyses as previously described.Citation30
Functional connectivity analysis
Ingenuity Pathway Analysis (IPA; Qiagen, Redwood City) was performed as previously describedCitation9 for analyzing, integrating, and interpreting the data generated from the methylation array.
Association with the recurrence-free survival of PCa patients
RNAseq data (RNAseqV2, level 3) from 497 PCa subjects as well as their associated clinical data (Biotab) were downloaded from the TCGA database on Feb 1st 2016. Expression levels of the 7 genes were pulled from normalized RNAseq gene results. The data were variance stabilizing transformed before dichotomizing into 2 groups by K-means clustering analysis. Recurrence-free survival was determined based on “days to new tumor event after initial treatment.” Survival analysis with log rank test as well as hazard ratio were calculated using GraphPad Prism software (La Jolla, CA).
Statistical analysis
Data are expressed as mean ± SEM. There were at least 3 individual experimental sets for gene expression analysis. Bisulfite sequencing (BS) analysis of the 7 selected gene candidates was performed on 6 individual samples, with ∼8–12 clones per group. One-way ANOVA and Tukey test were used for comparison among groups. P < 0.05 was considered as statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KEPI_A_1208891_s02.zip
Download Zip (106.2 KB)Acknowledgments
The results published here are in whole or in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. We thank The Genomics, Epigenomics, and Sequencing Core for the array service. We also thank Lynn Birch, Wen Yang Hu, Saikumar Karyala, Miral Patel, Hong Xiao and for their technical assistance and Jennifer Veevers for her excellent editing of the manuscript.
Funding
This study was supported in part by grants from the National Institutes of Environmental Health Sciences: R01CA015776 (SMH), R01ES015584 (GSP, SMH), RC2ES018758 (GSP, SMH, MM), RC2ES018789 (SMH, MM), U01ES019480 (SMH, MM), U01ES020988 (SMH, MM), CA172220 (GSP), and P30ES006096 (SMH, MM), the United States Department of Veterans Affairs I01BX000675 (SMH), and the Department of Defense Prostate Cancer Research Program W81XWH-06-1-0373 (WT) and W81XWH-15-1-0496 (AC).
References
- Rubin BS, Bisphenol A. an endocrine disruptor with widespread exposure and multiple effects. J Steroid Biochem Mol Biol 2011; 127:27-34; PMID:21605673; http://dx.doi.org/10.1016/j.jsbmb.2011.05.002
- Seachrist DD, Bonk KW, Ho SM, Prins GS, Soto AM, Keri RA. A review of the carcinogenic potential of bisphenol A. Reprod Toxicol 2015; 59:167-82; PMID:26493093; http://dx.doi.org/10.1016/j.reprotox.2015.09.006
- Vandenberg LN, Maffini MV, Sonnenschein C, Rubin BS, Soto AM. Bisphenol-A and the great divide: a review of controversies in the field of endocrine disruption. EndocrRev 2009; 30:75-95; PMID:19074586; http://dx.doi.org/10.1210/er.2008-0021
- Cavallaro M, Mariani J, Lancini C, Latorre E, Caccia R, Gullo F, Valotta M, DeBiasi S, Spinardi L, Ronchi A, et al. Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development 2008; 135:541-57; PMID:18171687; http://dx.doi.org/10.1242/dev.010801
- Chen K, Fallen S, Abaan HO, Hayran M, Gonzalez C, Wodajo F, MacDonald T, Toretsky JA, Uren A. Wnt10b induces chemotaxis of osteosarcoma and correlates with reduced survival. Pediatric blood & cancer 2008; 51:349-55; PMID:18465804; http://dx.doi.org/10.1002/pbc.21595
- Modugno F, Weissfeld JL, Trump DL, Zmuda JM, Shea P, Cauley JA, Ferrell RE. Allelic variants of aromatase and the androgen and estrogen receptors: toward a multigenic model of prostate cancer risk. Clin Cancer Res 2001; 7:3092-6; PMID:11595700
- Otsubo T, Akiyama Y, Yanagihara K, Yuasa Y. SOX2 is frequently downregulated in gastric cancers and inhibits cell growth through cell-cycle arrest and apoptosis. Br J Cancer 2008; 98:824-31; PMID:18268498; http://dx.doi.org/10.1038/sj.bjc.6604193
- Calafat AM, Kuklenyik Z, Reidy JA, Caudill SP, Ekong J, Needham LL. Urinary concentrations of bisphenol A and 4-nonylphenol in a human reference population. Environ Health Perspect 2005; 113:391-5; PMID:15811827; http://dx.doi.org/10.1289/ehp.7534
- Lam HM, Ho SM, Chen J, Medvedovic M, Tam NN. Bisphenol A Disrupts HNF4alpha-Regulated Gene Networks Linking to Prostate Preneoplasia and Immune Disruption in Noble Rats. Endocrinology 2016; 157:207-19; PMID:26496021; http://dx.doi.org/10.1210/en.2015-1363
- Tarapore P, Ying J, Ouyang B, Burke B, Bracken B, Ho SM. Exposure to bisphenol A correlates with early-onset prostate cancer and promotes centrosome amplification and anchorage-independent growth in vitro. PloS one 2014; 9:e90332; PMID:24594937; http://dx.doi.org/10.1371/journal.pone.0090332
- Balakrishnan B, Henare K, Thorstensen EB, Ponnampalam AP, Mitchell MD. Transfer of bisphenol A across the human placenta. AmJObstetGynecol 2010; 202:393-7; PMID:20350650; http://dx.doi.org/10.1016/j.ajog.2010.01.025
- Gerona RR, Woodruff TJ, Dickenson CA, Pan J, Schwartz JM, Sen S, Friesen MW, Fujimoto VY, Hunt PA. Bisphenol-A (BPA), BPA glucuronide, and BPA sulfate in midgestation umbilical cord serum in a northern and central California population. Environ Sci Technol 2013; 47:12477-85; PMID:23941471; http://dx.doi.org/10.1021/es402764d
- Lee YJ, Ryu HY, Kim HK, Min CS, Lee JH, Kim E, Nam BH, Park JH, Jung JY, Jang DD, et al. Maternal and fetal exposure to bisphenol A in Korea. ReprodToxicol 2008; 25:413-9; PMID:18577445; http://dx.doi.org/10.1016/j.reprotox.2008.05.058
- Padmanabhan V, Siefert K, Ransom S, Johnson T, Pinkerton J, Anderson L, Tao L, Kannan K. Maternal bisphenol-A levels at delivery: a looming problem? JPerinatol 2008; 28:258-63; PMID:18273031; http://dx.doi.org/10.1038/sj.jp.7211913
- Edginton AN, Ritter L. Predicting plasma concentrations of bisphenol A in children younger than 2 years of age after typical feeding schedules, using a physiologically based toxicokinetic model. Environ Health Perspect 2009; 117:645-52; PMID:19440506; http://dx.doi.org/10.1289/ehp.0800073
- Wetherill YB, Akingbemi BT, Kanno J, McLachlan JA, Nadal A, Sonnenschein C, Watson CS, Zoeller RT, Belcher SM. In vitro molecular mechanisms of bisphenol A action. Reprod Toxicol 2007; 24:178-98; PMID:17628395; http://dx.doi.org/10.1016/j.reprotox.2007.05.010
- Hu WY, Shi GB, Lam HM, Hu DP, Ho SM, Madueke IC, Kajdacsy-Balla A, Prins GS. Estrogen-initiated transformation of prostate epithelium derived from normal human prostate stem-progenitor cells. Endocrinology 2011; 152:2150-63; PMID:21427218; http://dx.doi.org/10.1210/en.2010-1377
- Lau KM, Leav I, Ho SM. Rat estrogen receptor-alpha and -beta, and progesterone receptor mRNA expression in various prostatic lobes and microdissected normal and dysplastic epithelial tissues of the Noble rats. Endocrinology 1998; 139:424-7; PMID:9421443; http://dx.doi.org/10.1210/endo.139.1.5809#sthash.rMzyrNHt.dpuf
- Prins GS, Hu WY, Shi GB, Hu DP, Majumdar S, Li G, Huang K, Nelles JL, Ho SM, Walker CL, et al. Bisphenol A promotes human prostate stem-progenitor cell self-renewal and increases in vivo carcinogenesis in human prostate epithelium. Endocrinology 2014; 155:805-17; PMID:24424067; http://dx.doi.org/10.1210/en.2013-1955
- Acconcia F, Pallottini V, Marino M. Molecular Mechanisms of Action of BPA. Dose-response 2015; 13:1559325815610582; PMID:26740804; http://dx.doi.org/10.1177/1559325815610582
- Moriyama K, Tagami T, Akamizu T, Usui T, Saijo M, Kanamoto N, Hataya Y, Shimatsu A, Kuzuya H, Nakao K. Thyroid hormone action is disrupted by bisphenol A as an antagonist. J Clin Endocrinol Metab 2002; 87:5185-90; PMID:12414890; http://dx.doi.org/10.1210/jc.2002-020209
- Lammoglia MA, Short RE, Bellows SE, Bellows RA, MacNeil MD, Hafs HD. Induced and synchronized estrus in cattle: dose titration of estradiol benzoate in peripubertal heifers and postpartum cows after treatment with an intravaginal progesterone-releasing insert and prostaglandin F2alpha. J Anim Sci 1998; 76:1662-70; PMID:9655587; http://dx.doi.org/10.2527/1998.7661662x
- Matthews J, Celius T, Halgren R, Zacharewski T. Differential estrogen receptor binding of estrogenic substances: a species comparison. J Steroid Biochem Mol Biol 2000; 74:223-34; PMID:11162928; http://dx.doi.org/10.1016/S0960-0760(00)00126-6
- Toyama Y, Yuasa S. Effects of neonatal administration of 17beta-estradiol, beta-estradiol 3-benzoate, or bisphenol A on mouse and rat spermatogenesis. Reprod Toxicol 2004; 19:181-8; PMID:15501383; http://dx.doi.org/10.1016/j.reprotox.2004.08.003
- Ho SM, Tang WY, Belmonte de FJ, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res 2006; 66:5624-32; PMID:16740699; http://dx.doi.org/10.1158/0008-5472.CAN-06-0516
- Prins GS, Birch L, Habermann H, Chang WY, Tebeau C, Putz O, Bieberich C. Influence of neonatal estrogens on rat prostate development. Reprod Fertil Dev 2001; 13:241-52; PMID:11800163; http://dx.doi.org/10.1071/RD00107
- Prins GS, Ho SM. Early-life estrogens and prostate cancer in an animal model. J Dev Orig Health Dis 2010; 1:365-70; PMID:24795802; http://dx.doi.org/10.1017/S2040174410000577
- Bernal AJ, Jirtle RL. Epigenomic disruption: the effects of early developmental exposures. Birth Defects Res A Clin Mol Teratol 2010; 88:938-44; PMID:20568270; http://dx.doi.org/10.1002/bdra.20685
- Bhan A, Hussain I, Ansari KI, Bobzean SA, Perrotti LI, Mandal SS. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol 2014; 141:160-70; PMID:24533973; http://dx.doi.org/10.1016/j.jsbmb.2014.02.002
- Ho SM, Cheong A, Lam HM, Hu WY, Shi GB, Zhu X, Chen J, Zhang X, Medvedovic M, Leung YK, et al. Exposure of Human Prostaspheres to Bisphenol A Epigenetically Regulates SNORD Family Noncoding RNAs via Histone Modification. Endocrinology 2015; 156:3984-95; PMID:26248216; http://dx.doi.org/10.1210/en.2015-1067
- Nahar MS, Liao C, Kannan K, Harris C, Dolinoy DC. In utero bisphenol A concentration, metabolism, and global DNA methylation across matched placenta, kidney, and liver in the human fetus. Chemosphere 2015; 124:54-60; PMID:25434263; http://dx.doi.org/10.1016/j.chemosphere.2014.10.071
- Tang WY, Morey LM, Cheung YY, Birch L, Prins GS, Ho SM. Neonatal exposure to estradiol/bisphenol A alters promoter methylation and expression of Nsbp1 and Hpcal1 genes and transcriptional programs of Dnmt3a/b and Mbd2/4 in the rat prostate gland throughout life. Endocrinology 2012; 153:42-55; PMID:22109888; http://dx.doi.org/10.1210/en.2011-1308
- Wong RL, Wang Q, Trevino LS, Bosland MC, Chen J, Medvedovic M, Prins GS, Kannan K, Ho SM, Walker CL. Identification of secretaglobin Scgb2a1 as a target for developmental reprogramming by BPA in the rat prostate. Epigenetics 2015; 10:127-34; PMID:25612011; http://dx.doi.org/10.1080/15592294.2015.1009768
- Prins GS, Ye SH, Birch L, Ho SM, Kannan K. Serum bisphenol A pharmacokinetics and prostate neoplastic responses following oral and subcutaneous exposures in neonatal Sprague-Dawley rats. Reprod Toxicol 2011; 31:1-9; PMID:20887781; http://dx.doi.org/10.1016/j.reprotox.2010.09.009
- Vermeulen A, Kaufman JM, Goemaere S, van Pottelberg I. Estradiol in elderly men. Aging Male 2002; 5:98-102; PMID:12198740; http://dx.doi.org/10.1080/tam.5.2.98.102
- Walker CL, Ho SM. Developmental reprogramming of cancer susceptibility. Nat Rev Cancer 2012; 12:479-86; PMID:22695395; http://dx.doi.org/10.1038/nrc3220
- Li LC. Designing PCR primer for DNA methylation mapping. Methods Mol Biol 2007; 402:371-84; PMID:17951806; http://dx.doi.org/10.1007/978-1-59745-528-2
- Chang SM, Chung LW. Interaction between prostatic fibroblast and epithelial cells in culture: role of androgen. Endocrinology 1989; 125:2719-27; PMID:2792005; http://dx.doi.org/10.1210/endo-125-5-2719
- Ho SM, Leav I, Damassa D, Kwan PW, Merk FB, Seto HS. Testosterone-mediated increase in 5 alpha-dihydrotestosterone content, nuclear androgen receptor levels, and cell division in an androgen-independent prostate carcinoma of Noble rats. Cancer Res 1988; 48:609-14; PMID:3257169
- Albertsen PC, Hanley JA, Fine J. 20-year outcomes following conservative management of clinically localized prostate cancer. Jama 2005; 293:2095-101; PMID:15870412; http://dx.doi.org/10.1001/jama.293.17.2095
- Prins GS, Huang L, Birch L, Pu Y. The role of estrogens in normal and abnormal development of the prostate gland. Ann N Y Acad Sci 2006; 1089:1-13; PMID:17261752; http://dx.doi.org/10.1196/annals.1386.009
- Ghosh PM, Malik SN, Bedolla RG, Wang Y, Mikhailova M, Prihoda TJ, Troyer DA, Kreisberg JI. Signal transduction pathways in androgen-dependent and -independent prostate cancer cell proliferation. Endocrine-related Cancer 2005; 12:119-34; PMID:15788644; http://dx.doi.org/10.1677/erc.1.00835
- Liao Y, Grobholz R, Abel U, Trojan L, Michel MS, Angel P, Mayer D. Increase of AKT/PKB expression correlates with gleason pattern in human prostate cancer. Int J Cancer 2003; 107:676-80; PMID:14520710; http://dx.doi.org/10.1002/ijc.11471
- Sanjeev S, MacLennan GT, Hartman DJ, Fu P, Resnick MI, and Gupta S. Activation of PI3K-Akt signaling pathway promotes prostate cancer cell invasion. International Journal of Cancer 2007; 121(7):1424-1432; PMID:17551921; http://dx.doi.org/10.1002/ijc.22862
- Lin HK, Yeh S, Kang HY, Chang C. Akt suppresses androgen-induced apoptosis by phosphorylating and inhibiting androgen receptor. Proc Natl Acad Sci U S A 2001; 98:7200-5; PMID:11404460; http://dx.doi.org/10.1073/pnas.121173298
- Price DT, Della Rocca G, Guo C, Ballo MS, Schwinn DA, Luttrell LM. Activation of extracellular signal-regulated kinase in human prostate cancer. J Urol 1999; 162:1537-42; PMID:10492251; http://dx.doi.org/10.1016/S0022-5347(05)68354-1
- Gao H, Ouyang X, Banach-Petrosky WA, Gerald WL, Shen MM, Abate-Shen C. Combinatorial activities of Akt and B-Raf/Erk signaling in a mouse model of androgen-independent prostate cancer. Proc Natl Acad Sci U S A 2006; 103:14477-82; PMID:16973750; http://dx.doi.org/10.1073/pnas.0606836103
- Jia X, Li X, Xu Y, Zhang S, Mou W, Liu Y, Liu Y, Lv D, Liu CH, Tan X, et al. SOX2 promotes tumorigenesis and increases the anti-apoptotic property of human prostate cancer cell. J Mol Cell Biol 2011; 3:230-8; PMID:21415100; http://dx.doi.org/10.1093/jmcb/mjr002
- Kregel S, Kiriluk KJ, Rosen AM, Cai Y, Reyes EE, Otto KB, Tom W, Paner GP, Szmulewitz RZ, Vander Griend DJ. Sox2 is an androgen receptor-repressed gene that promotes castration-resistant prostate cancer. PloS one 2013; 8:e53701; PMID:23326489; http://dx.doi.org/10.1371/journal.pone.0053701
- Miranda-Carboni GA, Krum SA, Yee K, Nava M, Deng QE, Pervin S, Collado-Hidalgo A, Galic Z, Zack JA, Nakayama K, et al. A functional link between Wnt signaling and SKP2-independent p27 turnover in mammary tumors. Gen Dev 2008; 22:3121-34; PMID:19056892; http://dx.doi.org/10.1101/gad.1692808
- Li X, Xu Y, Chen Y, Chen S, Jia X, Sun T, Liu Y, Li X, Xiang R, Li N. SOX2 promotes tumor metastasis by stimulating epithelial-to-mesenchymal transition via regulation of WNT/beta-catenin signal network. Cancer Lett 2013; 336:379-89; PMID:23545177; http://dx.doi.org/10.1016/j.canlet.2013.03.027
- Ummanni R, Teller S, Junker H, Zimmermann U, Venz S, Scharf C, Giebel J, Walther R. Altered expression of tumor protein D52 regulates apoptosis and migration of prostate cancer cells. FEBS J 2008; 275:5703-13; PMID:18959755; http://dx.doi.org/10.1111/j.1742-4658.2008.06697.x
- Tennstedt P, Bolch C, Strobel G, Minner S, Burkhardt L, Grob T, Masser S, Sauter G, Schlomm T, Simon R. Patterns of TPD52 overexpression in multiple human solid tumor types analyzed by quantitative PCR. Int J Oncol 2014; 44:609-15; PMID:24317684; http://dx.doi.org/10.3892/ijo.2013.2200
- van Duin M, van Marion R, Vissers K, Watson JE, van Weerden WM, Schroder FH, Hop WC, van der Kwast TH, Collins C, van Dekken H. High-resolution array comparative genomic hybridization of chromosome arm 8q: evaluation of genetic progression markers for prostate cancer. Genes Chromosomes Cancer 2005; 44:438-49; PMID:16130124; http://dx.doi.org/10.1002/gcc.20259
- Qi H, Fillion C, Labrie Y, Grenier J, Fournier A, Berger L, El-Alfy M, Labrie C. AIbZIP, a novel bZIP gene located on chromosome 1q21.3 that is highly expressed in prostate tumors and of which the expression is up-regulated by androgens in LNCaP human prostate cancer cells. Cancer Res 2002; 62:721-33; PMID:11830526
- Schmidt U, Fuessel S, Koch R, Baretton GB, Lohse A, Tomasetti S, Unversucht S, Froehner M, Wirth MP, Meye A. Quantitative multi-gene expression profiling of primary prostate cancer. Prostate 2006; 66:1521-34; PMID:16921506; http://dx.doi.org/10.1002/pros.20490
- Wissmann C, Wild PJ, Kaiser S, Roepcke S, Stoehr R, Woenckhaus M, Kristiansen G, Hsieh JC, Hofstaedter F, Hartmann A, et al. WIF1, a component of the Wnt pathway, is down-regulated in prostate, breast, lung, and bladder cancer. J Pathol 2003; 201:204-12; PMID:14517837; http://dx.doi.org/10.1002/path.1449
- Prins GS, Calderon-Gierszal EL, Hu WY. Stem Cells as Hormone Targets That Lead to Increased Cancer Susceptibility. Endocrinology 2015; 156:3451-7; PMID:26241068; http://dx.doi.org/10.1210/en.2015-1357
- Kim JW, Kim ST, Turner AR, Young T, Smith S, Liu W, Lindberg J, Egevad L, Gronberg H, Isaacs WB, et al. Identification of new differentially methylated genes that have potential functional consequences in prostate cancer. PloS one 2012; 7:e48455; PMID:23119026; http://dx.doi.org/10.1371/journal.pone.0048455
- Kato H, Araki T, Itoyama Y, Kogure K, Kato K. An immunohistochemical study of heat shock protein-27 in the hippocampus in a gerbil model of cerebral ischemia and ischemic tolerance. Neuroscience 1995; 68:65-71; PMID:7477936; http://dx.doi.org/10.1016/0306-4522(95)00141-5
- Alonso-Magdalena P, Ropero AB, Soriano S, Garcia-Arevalo M, Ripoll C, Fuentes E, Quesada I, Nadal A. Bisphenol-A acts as a potent estrogen via non-classical estrogen triggered pathways. Mol Cell Endocrinol 2012; 355:201-7; PMID:22227557; http://dx.doi.org/10.1016/j.mce.2011.12.012
- Chan QK, Lam HM, Ng CF, Lee AY, Chan ES, Ng HK, Ho SM, Lau KM. Activation of GPR30 inhibits the growth of prostate cancer cells through sustained activation of Erk1/2, c-jun/c-fos-dependent upregulation of p21, and induction of G(2) cell-cycle arrest. Cell Death Differ 2010; 17:1511-23; PMID:20203690; http://dx.doi.org/10.1038/cdd.2010.20
- Krishnan K, Gagne M, Nong A, Aylward LL, Hays SM. Biomonitoring Equivalents for bisphenol A (BPA). Regul Toxicol Pharmacol 2010; 58:18-24; PMID:20541576; http://dx.doi.org/10.1016/j.yrtph.2010.06.005
- Prins GS. Neonatal estrogen exposure induces lobe-specific alterations in adult rat prostate androgen receptor expression. Endocrinology 1992; 130:3703-14; PMID:1597166; http://dx.doi.org/10.1210/en.130.6.3703
- Draganov DI, Markham DA, Beyer D, Waechter JM, Jr., Dimond SS, Budinsky RA, Shiotsuka RN, Snyder SA, Ehman KD, Hentges SG. Extensive metabolism and route-dependent pharmacokinetics of bisphenol A (BPA) in neonatal mice following oral or subcutaneous administration. Toxicology 2015; 333:168-78; PMID:25929835; http://dx.doi.org/10.1016/j.tox.2015.04.012
- Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res 2006; 66:7939-47; PMID:16912168; http://dx.doi.org/10.1158/0008-5472.CAN-06-1888
- Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PloS one 2010; 5:e13100; PMID:20927350; http://dx.doi.org/10.1371/journal.pone.0013100
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Gen Biol 2004; 5:R80; PMID:15461798; http://dx.doi.org/10.1186/gb-2004-5-10-r80
- Newton MA, Quintana FA, den Boon JA. Random set methods identify distinct aspects of the enrichment signal in gene-set analysis. AnnApplStat 2007; 1:85-106; http://dx.doi.org/doi:10.1214/07-AOAS104
- Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics 2005; 21:4067-8; PMID:16141249; http://dx.doi.org/10.1093/bioinformatics/bti652
- van Belkum A, Melles DC, Nouwen J, van Leeuwen WB, van Wamel W, Vos MC, Wertheim HF, Verbrugh HA. Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect Genet Evol 2009; 9:32-47; PMID:19000784; http://dx.doi.org/10.1016/j.meegid.2008.09.012
- Liu H, Wei L, Tao Q, Deng H, Ming M, Xu P, Le W. Decreased NURR1 and PITX3 gene expression in Chinese patients with Parkinson's disease. Eur J Neurol 2012; 19:870-5; PMID:22309633; http://dx.doi.org/10.1111/j.1468-1331.2011.03644.x
- Lei Z, Jiang Y, Li T, Zhu J, Zeng S. Signaling of glial cell line-derived neurotrophic factor and its receptor GFRalpha1 induce Nurr1 and Pitx3 to promote survival of grafted midbrain-derived neural stem cells in a rat model of Parkinson disease. J Neuropathol Exp Neurol 2011; 70:736-47; PMID:21865882; http://dx.doi.org/10.1097/NEN.0b013e31822830e5
- Chen H, Wang Y, Xue F. Expression and the clinical significance of Wnt10a and Wnt10b in endometrial cancer are associated with the Wnt/beta-catenin pathway. Oncol Rep 2013; 29:507-14; PMID:23135473; http://dx.doi.org/10.3892/or.2012.2126
- Ouji Y, Nakamura-Uchiyama F, Yoshikawa M. Canonical Wnts, specifically Wnt-10b, show ability to maintain dermal papilla cells. Biochem Biophys Res Commun 2013; 438:493-9; PMID:23916705; http://dx.doi.org/10.1016/j.bbrc.2013.07.108
- Wend P, Runke S, Wend K, Anchondo B, Yesayan M, Jardon M, Hardie N, Loddenkemper C, Ulasov I, Lesniak MS, et al. WNT10B/beta-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO Mol Med 2013; 5:264-79; PMID:23307470; http://dx.doi.org/10.1002/emmm.201201320
- Thiele S, Rauner M, Goettsch C, Rachner TD, Benad P, Fuessel S, Erdmann K, Hamann C, Baretton GB, Wirth MP, et al. Expression profile of WNT molecules in prostate cancer and its regulation by aminobisphosphonates. J Cell Biochem 2011; 112:1593-600; PMID:21344486; http://dx.doi.org/10.1002/jcb.23070
- Calderon-Gierszal EL, Prins GS. Directed Differentiation of Human Embryonic Stem Cells into Prostate Organoids In Vitro and its Perturbation by Low-Dose Bisphenol A Exposure. PloS one 2015; 10:e0133238; PMID:26222054; http://dx.doi.org/10.1371/journal.pone.0133238
- Golestaneh N, Beauchamp E, Fallen S, Kokkinaki M, Uren A, Dym M. Wnt signaling promotes proliferation and stemness regulation of spermatogonial stem/progenitor cells. Reproduction 2009; 138:151-62; PMID:19419993; http://dx.doi.org/10.1530/REP-08-0510
- Gonez LJ, Naselli G, Banakh I, Niwa H, Harrison LC. Pancreatic expression and mitochondrial localization of the progestin-adipoQ receptor PAQR10. Mol Med 2008; 14:697-704; PMID:18769639; http://dx.doi.org/10.2119/2008-00072.Gonez
- Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 2014; 511:246-50; PMID:24909994; http://dx.doi.org/10.1038/nature13305
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron 2003; 39:749-65; PMID:12948443; http://dx.doi.org/10.1016/S0896-6273(03)00497-5
- Schoenhals M, Kassambara A, De Vos J, Hose D, Moreaux J, Klein B. Embryonic stem cell markers expression in cancers. Biochem Biophys Res Commun 2009; 383:157-62; PMID:19268426; http://dx.doi.org/10.1016/j.bbrc.2009.02.156
- Fernandez-Vega I, Garcia-Suarez O, Garcia B, Crespo A, Astudillo A, Quiros LM. Heparan sulfate proteoglycans undergo differential expression alterations in right sided colorectal cancer, depending on their metastatic character. BMC Cancer 2015; 15:742; PMID:26482785; http://dx.doi.org/10.1186/s12885-015-1724-9
- Lu Y, Futtner C, Rock JR, Xu X, Whitworth W, Hogan BL, Onaitis MW. Evidence that SOX2 overexpression is oncogenic in the lung. PloS one 2010; 5:e11022; PMID:20548776; http://dx.doi.org/10.1371/journal.pone.0011022
- Luo W, Li S, Peng B, Ye Y, Deng X, Yao K. Embryonic stem cells markers SOX2, OCT4 and Nanog expression and their correlations with epithelial-mesenchymal transition in nasopharyngeal carcinoma. PloS one 2013; 8:e56324; PMID:23424657; http://dx.doi.org/10.1371/journal.pone.0056324
- Rodriguez-Pinilla SM, Sarrio D, Moreno-Bueno G, Rodriguez-Gil Y, Martinez MA, Hernandez L, Hardisson D, Reis-Filho JS, Palacios J. Sox2: a possible driver of the basal-like phenotype in sporadic breast cancer. Modern Pathol 2007; 20:474-81; PMID:17334350; http://dx.doi.org/10.1038/modpathol.3800760
- Kopp JL, Ormsbee BD, Desler M, Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem cells 2008; 26:903-11; PMID:18238855; http://dx.doi.org/10.1634/stemcells.2007-0951
- Seo E, Basu-Roy U, Zavadil J, Basilico C, Mansukhani A. Distinct functions of Sox2 control self-renewal and differentiation in the osteoblast lineage. Mol Cell Biol 2011; 31:4593-608; PMID:21930787; http://dx.doi.org/10.1128/MCB.05798-11
- Wang R, He H, Sun X, Xu J, Marshall FF, Zhau H, Chung LW, Fu H, He D. Transcription variants of the prostate-specific PrLZ gene and their interaction with 14-3-3 proteins. Biochem Biophys Res Commun 2009; 389:455-60; PMID:19732746; http://dx.doi.org/10.1016/j.bbrc.2009.08.165
- Bian S, Akyuz N, Bernreuther C, Loers G, Laczynska E, Jakovcevski I, Schachner M. Dermatan sulfotransferase Chst14/D4st1, but not chondroitin sulfotransferase Chst11/C4st1, regulates proliferation and neurogenesis of neural progenitor cells. J Cell Sci 2011; 124:4051-63; PMID:22159417; http://dx.doi.org/10.1242/jcs.088120
- Lewis JD, Payton LA, Whitford JG, Byrne JA, Smith DI, Yang L, Bright RK. Induction of tumorigenesis and metastasis by the murine orthologue of tumor protein D52. Mol Cancer Res 2007; 5:133-44; PMID:17314271; http://dx.doi.org/10.1158/1541-7786.MCR-06-0245
- Wang R, Xu J, Saramaki O, Visakorpi T, Sutherland WM, Zhou J, Sen B, Lim SD, Mabjeesh N, Amin M, et al. PrLZ, a novel prostate-specific and androgen-responsive gene of the TPD52 family, amplified in chromosome 8q21.1 and overexpressed in human prostate cancer. Cancer Res 2004; 64:1589-94; PMID:14996714; http://dx.doi.org/10.1158/0008-5472.CAN-03-3331
- Ross AE, Marchionni L, Vuica-Ross M, Cheadle C, Fan J, Berman DM, Schaeffer EM. Gene expression pathways of high grade localized prostate cancer. Prostate 2011; 71:1568-77; PMID:21360566; http://dx.doi.org/10.1002/pros.21373
- Bismar TA, Demichelis F, Riva A, Kim R, Varambally S, He L, Kutok J, Aster JC, Tang J, Kuefer R, et al. Defining aggressive prostate cancer using a 12-gene model. Neoplasia 2006; 8:59-68; PMID:16533427; http://dx.doi.org/10.1593/neo.05664
- Byrne JA, Balleine RL, Schoenberg Fejzo M, Mercieca J, Chiew YE, Livnat Y, St Heaps L, Peters GB, Byth K, Karlan BY, et al. Tumor protein D52 (TPD52) is overexpressed and a gene amplification target in ovarian cancer. Int J Cancer 2005; 117:1049-54; PMID:15986428; http://dx.doi.org/10.1002/ijc.21250
- Alagaratnam S, Hardy JR, Lothe RA, Skotheim RI, Byrne JA. TPD52, a candidate gene from genomic studies, is overexpressed in testicular germ cell tumours. Mol Cell Endocrinol 2009; 306:75-80; PMID:19041365; http://dx.doi.org/10.1016/j.mce.2008.10.043
- Takeda T, Tezuka Y, Horiuchi M, Hosono K, Iida K, Hatakeyama D, Miyaki S, Kunisada T, Shibata T, Tezuka K. Characterization of dental pulp stem cells of human tooth germs. J Dental Res 2008; 87:676-81; PMID:18573990; http://dx.doi.org/10.1177/154405910808700716
- Ben Aicha S, Lessard J, Pelletier M, Fournier A, Calvo E, Labrie C. Transcriptional profiling of genes that are regulated by the endoplasmic reticulum-bound transcription factor AIbZIP/CREB3L4 in prostate cells. Physiol Genomics 2007; 31:295-305; PMID:17712038; http://dx.doi.org/10.1152/physiolgenomics.00097.2007
- Adham IM, Eck TJ, Mierau K, Muller N, Sallam MA, Paprotta I, Schubert S, Hoyer-Fender S, Engel W. Reduction of spermatogenesis but not fertility in Creb3l4-deficient mice. Mol Cell Biol 2005; 25:7657-64; PMID:16107712; http://dx.doi.org/10.1128/MCB.25.17.7657-7664.2005
