Abstract
Objectives
Previous results demonstrated that CYP2D6 and CYP2C19 gene variants affect serum concentrations of antidepressants. We implemented a PGx service determining gene variants in CYP2D6 and CYP2C19 in our clinical routine care and report on our first patient cohort.
Methods
We analysed CYP2D6 and CYP2C19 allele, genotype, and phenotype frequencies, and actionable pharmacogenetic variants in this German psychiatric inpatient cohort. Two-tailed z-test was used to investigate for differences in CYP2D6 and CYP2C19 phenotypes and actionable/non-actionable genetic variant frequencies between our cohort and reference cohorts.
Results
Out of the 154 patients included, 44.8% of patients were classified as CYP2D6 normal metabolizer, 38.3% as intermediate metabolizers, 8.4% as poor metabolizers, and 2.6% as ultrarapid metabolizers. As for CYP2C19, 40.9% of patients were classified as normal metabolizers, 19.5% as intermediate metabolizers, 2.6% as poor metabolizers, 31.2% as rapid metabolizers, and 5.8% as ultrarapid metabolizers. Approximately, 80% of patients had at least one actionable PGx variant.
Conclusion
There is a high prevalence of actionable PGx variants in psychiatric inpatients which may affect treatment response. Physicians should refer to PGx-informed dosing guidelines in carriers of these variants. Pre-emptive PGx testing in general may facilitate precision medicine also for other drugs metabolised by CYP2D6 and/or CYP2C19.
Introduction
Mood and anxiety are among the most prevalent mental health conditions globally, with a lifetime prevalence of 5%–10% according to the WHO (WHO Citation2023). In terms of depressive disorders, while there are effective pharmacological treatment interventions (Cipriani et al. Citation2018; Malhi and Mann Citation2018), up to two thirds of the patients do not respond to the initial administered antidepressant and about one third fail to respond following treatment approaches (Malhi and Mann Citation2018). Moreover, in more than one fourth of the patients taking antidepressants adverse drug reactions (ADR) occur (Mishra et al. Citation2013). Consequently, in psychiatry, strategies are warranted to provide the most effective treatment options for an individual patient, i.e. maximising the likelihood for response and minimise risk for ADRs (Menke Citation2018). The concept of precision medicine has received much attention, which aims to combine information gained from genetic markers, biomarkers, behavioural measurements, neuropsychological testing, imaging and computational science to provide a tailored antidepressant treatment approach to optimise treatment (Menke Citation2018; Menke et al. Citation2020).
In our pilot study at the University Hospital of Wuerzburg, our combined approach of using genetic information, therapeutic drug monitoring and clinical data to tailor antidepressant treatment (Menke et al. Citation2020), demonstrated that CYP2D6 and CYP2C19 gene variants affect serum concentrations of antidepressants (Scherf-Clavel et al. Citation2022). Our findings are consistent with existing clinical guidelines using genetic information of CYP2D6 and CYP2C19 for dose adjustments in tricyclic antidepressants, selective serotonin reuptake inhibitors, venlafaxine and antipsychotic medications (Hicks et al. Citation2015, Citation2017; Beunk et al. Citation2023; Bousman et al. Citation2023) provided by the Clinical Pharmacogenetics Implementation Consortium (CPIC) (Relling and Klein Citation2011; Relling et al. Citation2020) and the Dutch Pharmacogenetics Working Group (DPWG) (Swen et al. Citation2008, Citation2011). In addition, dosing recommendations for drugs used in psychiatry are also available for CYP2D6 and atomoxetine, and opioids (Brown et al. Citation2019; Crews et al. Citation2021). Consequently, also an expert group of the International Society of Psychiatric Genetics recommended the use of pharmacogenetic (PGx) testing for CYP2D6, and CYP2C19 (Bousman, Bengesser, et al. Citation2021). Reviewing existing literature, Murphy et al. also summarised, that for a large number of antidepressants and antipsychotics CYP2D6 and CYP2C19 genotype information should be used for treatment recommendations (Murphy et al. Citation2022).
In order to advance the field of precision medicine for the treatment of psychiatric diseases, we implemented a PGx testing service for determining CYP2D6 and CYP2C19 variants in clinical routine care at our Department of Psychiatry, Psychosomatics and Psychotherapy at the University Hospital of Wuerzburg.
In the first cohort of 154 patients, we present frequencies of the CYP2D6 and CYP2C19 alleles, genotypes, and respective phenotypes, and frequencies of actionable pharmacogenetic variants.
Methods
Patients
A total of 154 inpatients where genotype data was available were included in our analyses. The sample comprised only patients older than 18 years. Clinical diagnoses were obtained through a validated, standardised interview according to DSM-IV criteria (Structured Clinical Interview for DSM, SCID-I, First and Gibbon Citation2004) and recorded with complete medication lists. Genotypes of CYP2D6 and CYP2C19 were determined between March 2021 and March 2023 in patients referred for pharmacogenetic testing by their treating physicians.
Genotyping was performed as part of the clinical routine according to recommendations of the German Genetic Diagnostics Commission (Gendiagnostik-Kommission Citation2013, Citation2017) and using procedures outlined in the German Genetic Diagnostics Act with written informed consent obtained by each patient. The subsequent retrospective analysis of routine clinical data was conducted without additional explicit written informed but with consent provided by the Wuerzburg ethics committee (20230525 01) in accordance with the principles of the declaration of Helsinki.
Genotyping
Genotyping of CYP2D6 and CYP2C19 was performed at our institution using a MassArray Analyser 4 system (Agena Bioscience GmbH, Hamburg, Germany) based on a self-designed panel using SpectroCHIP®-96 Arrays and the iPLEX® Pro chemistry following the instructions supplied by the manufacturer. Primer sequences are available on request. Moreover, copy number variations (CNV) in CYP2D6 were determined using the CYP2D6 RealFast™ CNV Assay, provided by ViennaLab Diagnostics GmbH, Vienna, Austria (ViennaLabDiagnosticsGmbH 2021). This real-time PCR based method enables the detection of the total copies in a sample, but not the assignment of the individual copies to the respective alleles. Alleles identified included CYP2D6*1, *2, *3, *4, *6, *9, *10, *14, *17, *34, *35, *39, *41, *46, *58, *64, *69, *71, *82, *88, *114, and CYP2C19*1, *2, *3, *4, *8, *11, *17 (SNPs and star allele coverage; supplemental digital content 1). The *1 allele for CYP2D6 and CYP2C19 is not directly genotyped but is assigned to the “wild type” based on the absence of any variant in the panel for each gene, respectively. The laboratory was certified by a quality control program (INSTAND Citation2020). Haplotypes were defined for all analysed SNPs according to gene specific haplotype tables from the PharmVar homepage (https://www.pharmvar.org/genes; supplemental digital content 1). If SNP coverage was not clearly assignable to one haplotype due to phasing ascertainment, various possible haplotype options were given. Phenotypes of CYP2D6 and CYP2C19 were determined according to the Clinical Pharmacogenetics Implementation Consortium (CPIC) specifications (CPIC Citation2021).
Actionable PGx variants
Patients assigned a NM phenotype for CYP2D6, and CYP2C19 were defined as not having an actionable PGx variant, while patients assigned an IM, PM, RM, or UM phenotype were considered to have an actionable PGx variant. For these variants PGx-based dosing guidelines by CPIC and/or DPWG for selective serotonin reuptake inhibitors, tricyclic antidepressants, but also other medications have been published. These guidelines can be found on the PharmGKB website (https://www.pharmgkb.org/guidelineAnnotations; https://www.pharmgkb.org/gene/PA128/prescribingInfo#guideline-annotations; https://www.pharmgkb.org/gene/PA124/prescribingInfo#guideline-annotations; accessed 13/04/2023) (Whirl-Carrillo et al. Citation2012, Citation2021).
Statistical analyses
Statistical analyses were conducted in R v4.0.4 (RCoreTeam Citation2021). Chord plots were prepared using the “circlize” package (Gu et al. Citation2014).
Frequencies of allele, genotype, and phenotype frequencies were calculated.
Two-tailed z-test was used to investigate for differences in frequencies of CYP2D6 and CYP2C19 phenotypes and actionable/non-actionable genetic variants between cohorts. p < .05 was considered as significant. Only cohorts in which the way to assign phenotypes was in line with CPIC specifications were used.
Results
Patient sample
Within 2 years of routine PGx service, 154 inpatients were genotyped. Patients were on average 43.2 years old (±15.3 mean ± standard deviation (SD)), and 59.1% were female (N = 91).
Patients suffered from schizophrenia (F20, N = 7; 4.55%), bipolar effective disorder (F31; N = 28; 18.18%), depressive episodes (F32; N = 17; 9.74%), recurrent depressive disorder (F33; N = 92; 59.74%), other anxiety disorders (F41; N = 1; 0.6%), obsessive-compulsive disorder (F42; N = 2; 1.3%), eating disorder (F50, N = 1; 0.65%), psychological and behavioural disorders associated with sexual development and orientation (F66; N = 1; 0.65%), and hyperkinetic disorder (F90; N = 1; 0.65%). For details see supplemental digital content 2. Clinical data on treatment response were not available.
Patients received between 1 and 17 drugs (mean ± SD 4.6 ± 3.2). Focusing on psychiatric medication, patients received between 0 and 7 psychiatric drugs (mean ± SD 2.7 ± 1.3). Most of the patients received quetiapine (34.5%), followed by amitriptyline (28.4%), mirtazapine (23.6%), and venlafaxine (15.5%). Regarding non-psychiatric medication, patients received between 0 and 14 non-psychiatric concomitant medication (mean ± SD 1.9 ± 2.8). Detailed information on medication are provided in supplemental digital content 2.
Forty (=30%) and 26 (=20%) of the recorded drugs were metabolised at least with participation of CYP2D6, and CYP2C19, respectively. Patients received between 0 and 6 drugs with CYP2D6 participation in the metabolism (mean ± SD 2.1 ± 1.1), and between 0 and 5 drugs with CYP2C19 participation in the metabolism (mean ± SD 1.1 ± 0.9). Detailed information are provided in supplemental digital content 2. Information on the metabolic pathways of the drugs were derived from the PSIAC table (online database for querying of drug interactions; Hiemke et al. Citation2024).
Allele frequencies
The allele frequencies of CYP2D6 and CYP2C19 evaluated in our routine PGx service are summarised in supplemental digital content 3. For allele frequency calculations, 59 patients were excluded for CYP2D6 and one patient for CYP2C19 because the haplotypes could not be determined unambiguously due to several possible allele combinations. Occurring at frequencies of 33.2%, 18.4%, and 22.6%, the CYP2D6*1, CYP2D6*2, and CYP2D6*4 were the three most common CYP2D6 alleles. The most common variant allele for CYP2C19 was the increased function *17 allele with a frequency of 25.8%.
Genotype and phenotype frequencies
Genotype frequencies were calculated for both CYP2D6 and CYP2C19 and are summarised in supplemental digital content 3.
Sixty-nine patients (44.8%) showed a CYP2D6 normal metabolizer (NM) status, 59 (38.3%) were classified as intermediate metabolizers (IM), 13 (8.4%) as poor metabolizers (PM), and 3 (1.9%) as ultrarapid metabolizers (UM). In nine patients (5.8%), CYP2D6 phenotype could not be classified (; ). In two patients a genotype with unknown/uncertain function was determined, in three patients more than one possible genotype was determined, which were assigned to different phenotypes, in four patients allele-duplication was present, but, as we could not specify which allele was duplicated, we cannot determine the phenotype unambiguously. Frequencies of CYP2D6 phenotypes were not different to the biogeographical group of Europeans (PharmGKB Citation2023d) for NM (p = .68), IM (p = 1.0), PM (p = .81), and UM (p = 1.0; ). Frequencies of CYP2D6 activity scores are reported in supplemental digital content 3.
Figure 1. Phenotype frequencies of CYP2C19 (blue) and CYP2D6 (orange). Chord plot showing CYP2D6/CYP2C19 combination in patients. Only 20.7% were normal metabolizer in both enzymes (dotted line).
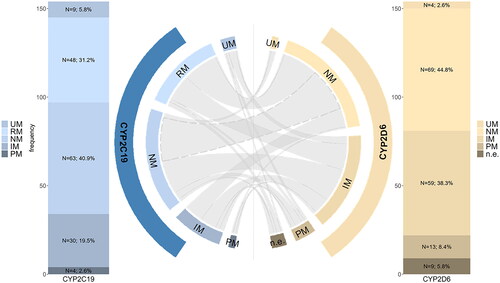
Table 1. Frequencies of CYP2D6 and CYP2C19 phenotypes.
As for CYP2C19, 63 patients (40.9%) were classified as NM, 30 (19.5%) as IM, 4 (2.6%) as PM, 48 (31.2%) as rapid metabolizers (RM), and 9 (5.8%) as UM (; ). Frequencies of CYP2C19 phenotypes were not significantly different to a biogeographical comparison group of Europeans (PharmGKB Citation2023c) for NM (p = 0.97), PM (p = 1.0), IM (p = 0.35), RM (p = 0.63), and UM (p = 0.95).
Actionable PGx variants for CYP2D6 and CYP2C19
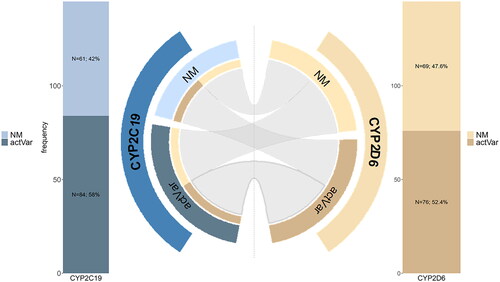
Considering 145 patients with complete data, only 30 patients (20.7%) did not carry an actionable PGx variant on CYP2D6 and CYP2C19. In contrast, 31 patients (21.4%) carried an actionable PGx variant only on CYP2D6, 39 (26.9%) exclusively on CYP2C19, and 45 patients (31.0%) on both genes ().
Figure 2. Frequencies of actionable gene variants in CYP2C19 (blue) and CYP2D6 (orange). Chord plot showing CYP2D6/CYP2C19 combination in patients. 45 patients (31.0%) carried an actionable PGx variant on both genes (bolded line). actVar; actionable Variant.
Frequencies of actionable/not actionable genotypes in CYP2D6 and CYP2C19 did not differ in different cohorts, with exception of the CYP2D6 phenotype frequencies of a study from Denmark (Lunenburg et al. Citation2021), and one large-scale genetic study from the USA (Li et al. Citation2023; ).
Table 2. Frequencies of not actionable/actionable gene variants in CYP2D6 and CYP2C19 found in different cohorts.
Discussion
Allele, genotype, and phenotype frequencies, as well as actionable PGx variants were calculated in psychiatric inpatients in a routine clinical setting.
Only 33.2% of the patients were carriers of CYP2D6*1 allele, with only 16.2% being carrier of the wild type (2 N*1/*1) for CYP2D6. Nevertheless, accounting for all patients with a CYP2D6 activity score of 1.25–2, 44.8% were classified as CYP2D6 NM. The frequency of patients with a CYP2D6 activity score of 2 is in accordance with the frequency in the European population (PharmGKB Citation2023d).
The CYP2C19*1 allele was present in 62.1% of the patients, with only 40.9% being carriers of the CYP2C19 wild type (*1/*1) and, therefore, being classified as CYP2C19 NM. CYP2C19 phenotype frequencies were in accordance with the frequency in the European population (PharmGKB Citation2023c).
With our results, we replicated previous data of German psychiatric inpatients (Hahn et al. Citation2021). Previously, genetic variants were determined using a commercially kit, including not only CYP2D6, and CYP2C19, but for example, also CYP3A4, CYP3A5, ABCB1, and COMT (Hahn et al. Citation2021). However, so far, for clinical routine, experts recommend that CYP2D6 and CYP2C19, and, in case of sertraline, CYP2B6 genotype information can be used in addition for treatment recommendations of psychiatric drugs (Bousman, Bengesser, et al. Citation2021; Murphy et al. Citation2022). Considered CYP2D6, and CYP2C19 in combination, previously, only 13% of the patients showed NM status for both enzymes (Hahn et al. Citation2021). This also is in accordance with our sample (20.7%) (two-tailed Z-test, p = .17). The limited number of NM on both enzymes support the relevance of panel-based testing compared to single-gene testing (Bousman C et al. Citation2019), as it is likely that multiple actionable PGx variants can be identified within one individual and enzymes with altered function possibly can be compensated by other enzymes involved in the metabolism of the drug. In our sample in four out of five patients, genetic variations may affect medication treatment, and, therefore, physicians should be educated and encouraged to use PGx-informed dosing guidelines provided by CPIC and DPWG (Swen et al. Citation2008, Citation2011; Relling and Klein Citation2011; Hicks et al. Citation2015; Citation2017; Relling et al. Citation2020; Beunk et al. Citation2023; Bousman et al. Citation2023). This is even more important as it was shown prior, that at admission, as well as at discharge from hospital, nearly in 1 of 2 patients, medication with an actionable PGx guideline recommendation were prescribed (Mostafa et al. Citation2021). In our sample, 102 patients were treated with medication with a CPIC PGx guideline recommendation (CPIC Citation2023b, Citation2023a). Moreover, 140 patients were treated with at least one drug with CYP2D6 participation in its metabolism, and 109 patients were treated with at least one drug with CYP2C19 participation in its metabolism. In addition, 101, and 42 patients were treated with two or more drugs with at least participation of CYP2D6, and CYP2C19, respectively, involved in the metabolism.
The clinical relevance of our findings might be affected by phenoconversion effects due to at least one concomitant medication potentially affecting CYP2D6 or CYP2C19. While phenoconversion is increasingly investigated and better understood (Bousman, Wu, et al. Citation2021; Mostafa et al. Citation2021; Scherf-Clavel et al. Citation2023), it is not yet known, how polypharmacy in addition to genetic variants affects functional enzyme status (Bousman, Wu, et al. Citation2021) including our sample. However, it may be likely that genetic variants in combination with polypharmacy may affect treatment of most of the patients in our sample. Phenoconversion effects would need to be addressed in future studies while some labs or commercial companies provide phenoconversion information for a given patient along with genetic information (Bousman, Wu, et al. Citation2021).
To date, in our hospital, due to the restrictions of the German Genetic Diagnostics Act, combined phenotype information of CYP2D6, and CYP2C19 are directly reported to the physician, who requested the PGx. This allows using the individual phenotype information for dosing recommendations provided by the CPIC (CPIC Citation2023a, Citation2023b) to personalise therapy during the current inpatient treatment. We are working on a direct transfer into the electronic patient file so that the information will be easily available for individualised therapy during future inpatient treatments.
Frequency of actionable genotypes either for CYP2D6, or CYP2C19 did not differ from reference populations (CPIC Citation2021; PharmGKB Citation2023c, Citation2023d), with exception of the CYP2D6 phenotype frequencies of a large population-based case-cohort study from Denmark (Lunenburg et al. Citation2021), and a study from Li et al. (Li et al. Citation2023) In these studies actionable CYP2D6 variants were less common than in our sample. However, Lunenburg et al. and Li et al. did not include CYP2D6 copy number variations in the analyses (Lunenburg et al. Citation2021; Li et al. Citation2023); therefore, CYP2D6 UM were not determined, and PM may be underestimated. High frequencies of actionable genotypes are not limited to psychiatric inpatients, but occur in the general population. In this context, PGx may not only be recommended in psychiatry, but pre-emptive PGx testing may be advantageous for each individual in order to facilitate uptake of precision medicine also for other important drugs metabolised by CYP2D6 and/or CYP2C19, for example, anticancer drugs, gastrointestinal, anti-inflammatory or cardiovascular agents (Bousman et al. Citation2019; PharmGKB Citation2023a, Citation2023b). Notably, dosing guidelines are available for many of these drugs as well (CPIC Citation2023a, Citation2023b).
Strengths and limitations
One major strength of our analysis has been the implementation of pharmacogenetic testing within a pre-existing clinical setting, allowing us to recruit a diagnostically homogeneous sample, representative for a German inpatient cohort affected with depressive symptoms. We have shown a notable and high proportion of patients are carriers of actionable genetic variants, relevant for psychiatric and non-psychiatric medications. However, there also are several limitations. Our sample size was limited, and genotyping was restricted to certain SNPs with known pharmacokinetic influence. Due to phasing ascertainment, some haplotypes could not be assigned to a specific phenotype and CNV determination did not allow clear assignment of the individual copies to a specific allele. Furthermore, clinical data on treatment response were not available, and potential phenoconversion effects were not assessed.
To overcome these methodical limitations, further studies in larger well described samples, with higher genetic resolution by sequencing methods and allele-specific CNV assignment using comparative genomic hybridisation (CGH) or specific algorithms (e.g. accurate genome-wide allele-specific copy number profiles (ASCAT) algorithm) on microarray or sequencing data, are necessary.
Conclusion
As a next and logical step in our efforts to advance precision medicine in psychiatry, we implemented a PGx service determining variants in CYP2D6 and CYP2C19 in clinical routine care at our institution. Routine data showed that only 44.8% of the patients were CYP2D6 normal metabolizers. Also, only 40.9% of the patients were CYP2C19 NM. Therefore, our study results indicate that there is a high prevalence (4 in 5 patients) of actionable genotypes in CYP2D6 and/or CYP2C19 in our psychiatric inpatients. In summary, PGx may not only be recommended in psychiatry, but pre-emptive PGx testing may be advantageous for all people in order to facilitate uptake of precision medicine also for other important drugs metabolised by CYP2D6 and/or CYP2C19. As such our result further support the need to broadly implement PGx testing in clinical routine.
Statement of interest
J. Deckert and H. Weber receive funding from the Deutschen Zentrum für Luft- und Raumfahrt (DLR) - Förderkennzeichen 01EK2204G (P4D, Project SP1, SP5A and Coordination). D. Müller has received research grants from CAN-BIND, Canadian Institutes of Health Research, Nubiyota, Ontario Brain Institute, CAMH AFP Innovation Funds and the CAMH Foundation. M. Scherf-Clavel, and S. Unterecker have no conflicts of interest.
Supplemental Material
Download MS Word (26.6 KB)Supplemental Material
Download MS Word (39.9 KB)Supplemental Material
Download MS Word (20.1 KB)Acknowledgements
None.
References
- Beunk L, Nijenhuis M, Soree B, de Boer-Veger NJ, Buunk AM, Guchelaar HJ, Houwink EJF, Risselada A, Rongen G, van Schaik RHN, et al. 2023. Dutch pharmacogenetics working group (DPWG) guideline for the gene-drug interaction between CYP2D6, CYP3A4 and CYP1A2 and antipsychotics. Eur J Hum Genet. 32(3):278–285. doi: 10.1038/s41431-023-01347-3.
- Bousman CA, Bengesser SA, Aitchison KJ, Amare AT, Aschauer H, Baune BT, Asl BB, Bishop JR, Burmeister M, Chaumette B, et al. 2021. Review and consensus on pharmacogenomic testing in psychiatry. Pharmacopsychiatry. 54(1):5–17. doi: 10.1055/a-1288-1061.
- Bousman C, Maruf AA, Müller DJ. 2019. Towards the integration of pharmacogenetics in psychiatry: a minimum, evidence-based genetic testing panel. Curr Opin Psychiatry. 32(1):7–15. doi: 10.1097/YCO.0000000000000465.
- Bousman CA, Stevenson JM, Ramsey LB, Sangkuhl K, Hicks JK, Strawn JR, Singh AB, Ruaño G, Mueller DJ, Tsermpini EE, et al. 2023. Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2D6, CYP2C19, CYP2B6, SLC6A4, and HTR2A genotypes and serotonin reuptake inhibitor antidepressants. Clin Pharmacol Ther. 114(1):51–68. doi: 10.1002/cpt.2903.
- Bousman CA, Wu P, Aitchison KJ, Cheng T. 2021. Sequence2Script: a web-based tool for translation of pharmacogenetic data into evidence-based prescribing recommendations. Front Pharmacol. 12:636650. doi: 10.3389/fphar.2021.636650.
- Bousman CA, Zierhut H, Müller DJ. 2019. Navigating the labyrinth of pharmacogenetic testing: a guide to test selection. Clin Pharmacol Ther. 106(2):309–312. doi: 10.1002/cpt.1432.
- Brown JT, Bishop JR, Sangkuhl K, Nurmi EL, Mueller DJ, Dinh JC, Gaedigk A, Klein TE, Caudle KE, McCracken JT, et al. 2019. Clinical pharmacogenetics implementation consortium guideline for cytochrome P450 (CYP)2D6 genotype and atomoxetine therapy. Clin Pharmacol Ther. 106(1):94–102. eng. doi: 10.1002/cpt.1409.
- Cipriani A, Furukawa TA, Salanti G, Chaimani A, Atkinson LZ, Ogawa Y, Leucht S, Ruhe HG, Turner EH, Higgins JPT, et al. 2018. Comparative efficacy and acceptability of 21 antidepressant drugs for the acute treatment of adults with major depressive disorder: a systematic review and network meta-analysis. Lancet. 391(10128):1357–1366. doi: 10.1016/S0140-6736(17)32802-7.
- CPIC. 2021. CPIC - Clinical Pharmacogenetics Implementation Consortium. [accessed 2022 March 8]. https://cpicpgx.org/.
- CPIC. 2023a. CYP2C19 CPIC guidelines. Clinical Pharmacogenetics Implementation Consortium; [accessed 2023 April 27]. https://cpicpgx.org/gene/cyp2c19/.
- CPIC. 2023b. CYP2D6 CPIC guidelines. Clinical Pharmacogenetics Implementation Consortium; [accessed 2023 April 27]. https://cpicpgx.org/gene/cyp2d6/.
- Crews KR, Monte AA, Huddart R, Caudle KE, Kharasch ED, Gaedigk A, Dunnenberger HM, Leeder JS, Callaghan JT, Samer CF, et al. 2021. Clinical pharmacogenetics implementation consortium guideline for CYP2D6, OPRM1, and COMT genotypes and select opioid therapy. Clin Pharmacol Ther. 110(4):888–896. doi: 10.1002/cpt.2149.
- First MB, Gibbon M. 2004. The structured clinical interview for DSM-IV axis I disorders (SCID-I) and the structured clinical interview for DSM-IV axis II disorders (SCID-II). Comprehensive handbook of psychological assessment, vol 2: personality assessment. Hoboken, NJ, US: John Wiley & Sons, Inc.; p. 134–143.
- Gendiagnostik-Kommission. 2013. Richtlinie der Gendiagnostik-Kommission (GEKO) für die anforderungen an die qualitätssicherung genetischer analysen zu medizinischen zwecken gemäß § 23 abs. 2 nr. 4 GenDG. Bundesgesundheitsblatt. 56:163–168.
- Gendiagnostik-Kommission. 2017. Richtlinie der Gendiagnostik-Kommission (GEKO) für die beurteilung genetischer eigenschaften hinsichtlich ihrer bedeutung für die wirkung eines arzneimittels bei einer behandlung gemäß §23 abs. 2 nr. 1b GenDG. Bundesgesundheitsblatt. 60:472–475.
- Gu Z, Gu L, Eils R, Schlesner M, Brors B. 2014. Circlize implements and enhances circular visualization in R. Bioinformatics. 30(19):2811–2812. doi: 10.1093/bioinformatics/btu393.
- Hahn M, Müller DJ, Roll SC. 2021. Frequencies of genetic polymorphisms of clinically relevant gene-drug pairs in a german psychiatric inpatient population. Pharmacopsychiatry. 54(2):81–89. doi: 10.1055/a-1312-7175.
- Hicks JK, Bishop JR, Sangkuhl K, Müller DJ, Ji Y, Leckband SG, Leeder JS, Graham RL, Chiulli DL, Ll A, et al. 2015. Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 98(2):127–134. doi: 10.1002/cpt.147.
- Hicks JK, Sangkuhl K, Swen JJ, Ellingrod VL, Müller DJ, Shimoda K, Bishop JR, Kharasch ED, Skaar TC, Gaedigk A, et al. 2017. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 102(1):37–44. doi: 10.1002/cpt.597.
- Hiemke C, Haen E, Eckermann G, Dobmeier M, Singer M, Silva M, Wenzel-Seifert K, Nagel-Hiemke M. 2024. PSIAC. Springer-Verlag GmbH, Heidelberg; [accessed 2019 March 18]. https://www.psiac.de/.
- INSTAND. 2020. INSTAND Gesellschaft zur Förderung der Qualitätssicherung in medizinischen Laboratorien e. V. [accessed 2020 Feb 10]. https://www.instand-ev.de/ueber-instand-ev/instand-ev.html.
- Li B, Sangkuhl K, Whaley R, Woon M, Keat K, Whirl-Carrillo M, Ritchie MD, Klein TE. 2023. Frequencies of pharmacogenomic alleles across biogeographic groups in a large-scale biobank. Am J Hum Genet. 110(10):1628–1647. doi: 10.1016/j.ajhg.2023.09.001.
- Lunenburg C, Thirstrup JP, Bybjerg-Grauholm J, Bækvad-Hansen M, Hougaard DM, Nordentoft M, Werge T, Børglum AD, Mors O, Mortensen PB, et al. 2021. Pharmacogenetic genotype and phenotype frequencies in a large danish population-based case-cohort sample. Transl Psychiatry. 11(1):294. doi: 10.1038/s41398-021-01417-4.
- Malhi GS, Mann JJ. 2018. Depression. Lancet. 392(10161):2299–2312. doi: 10.1016/S0140-6736(18)31948-2.
- Menke A. 2018. Precision pharmacotherapy: psychiatry’s future direction in preventing, diagnosing, and treating mental disorders. Pharmgenomics Pers Med. 11:211–222. doi: 10.2147/PGPM.S146110.
- Menke A, Weber H, Deckert J. 2020. Roadmap for routine pharmacogenetic testing in a psychiatric university hospital. Pharmacopsychiatry. 53(4):179–183. doi: 10.1055/a-0914-3234.
- Mishra S, Swain TR, Mohanty M. 2013. Adverse drug reaction monitoring of antidepressants in the psychiatry outpatients department of a tertiary care teaching hospital. J Clin Diagn Res. 7(6):1131–1134.
- Mostafa S, Polasek TM, Sheffield LJ, Huppert D, Kirkpatrick CMJ. 2021. Quantifying the impact of phenoconversion on medications with actionable pharmacogenomic guideline recommendations in an acute aged persons mental health setting. Front Psychiatry. 12:724170. doi: 10.3389/fpsyt.2021.724170.
- Murphy LE, Fonseka TM, Bousman CA, Müller DJ. 2022. Gene-drug pairings for antidepressants and antipsychotics: level of evidence and clinical application. Mol Psychiatry. 27(1):593–605. doi: 10.1038/s41380-021-01340-6.
- PharmGKB. 2023a. CYP2C19 pathways. PharmGKB. [accessed 2023 April 27]. https://www.pharmgkb.org/gene/PA124/pathway.
- PharmGKB. 2023b. CYP2D6 pathways. PharmGKB. [accessed 2023 April 27]. https://www.pharmgkb.org/gene/PA128/pathway.
- PharmGKB. 2023c. Gene-specific information tables for CYP2C19. PharmGKB. [accessed 2023 April 13]. https://www.pharmgkb.org/page/cyp2c19RefMaterials.
- PharmGKB. 2023d. Gene-specific information tables for CYP2D6. PharmGKB. [accessed 2023 April 13]. https://www.pharmgkb.org/page/cyp2d6RefMaterials.
- RCoreTeam. 2021. R: A language and environment for statistical computing. https://www.R-project.org. R Foundation for Statistical Computing, Vienna, Austria.
- Relling MV, Klein TE. 2011. CPIC: clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin Pharmacol Ther. 89(3):464–467. doi: 10.1038/clpt.2010.279.
- Relling MV, Klein TE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Caudle KE. 2020. The clinical pharmacogenetics implementation consortium: 10 years later. Clin Pharmacol Ther. 107(1):171–175. doi: 10.1002/cpt.1651.
- Scherf-Clavel M, Frantz A, Eckert A, Weber H, Unterecker S, Deckert J, Reif A, Hahn M. 2023. Effect of CYP2D6 pharmacogenetic phenotype and phenoconversion on serum concentrations of antidepressants and antipsychotics: a retrospective cohort study. Int J Clin Pharm. 45(5):1107–1117. doi: 10.1007/s11096-023-01588-8.
- Scherf-Clavel M, Weber H, Wurst C, Stonawski S, Hommers L, Unterecker S, Wolf C, Domschke K, Rost N, Brückl T, et al. 2022. Effects of pharmacokinetic gene variation on therapeutic drug levels and antidepressant treatment response. Pharmacopsychiatry. 55(5):246–254. doi: 10.1055/a-1872-0613.
- Swen JJ, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, Mulder H, Rongen GA, van Schaik RH, Schalekamp T, Touw DJ, et al. 2011. Pharmacogenetics: from bench to byte–an update of guidelines. Clin Pharmacol Ther. 89(5):662–673. doi: 10.1038/clpt.2011.34.
- Swen JJ, Wilting I, de Goede AL, Grandia L, Mulder H, Touw DJ, de Boer A, Conemans JM, Egberts TC, Klungel OH, et al. 2008. Pharmacogenetics: from bench to byte. Clin Pharmacol Ther. 83(5):781–787. doi: 10.1038/sj.clpt.6100507.
- ViennaLab Diagnostics GmbH. 2021. CYP2D6 RealFast CNV assay. ViennaLab Diagnostics GmbH.
- Whirl-Carrillo M, Huddart R, Gong L, Sangkuhl K, Thorn CF, Whaley R, Klein TE. 2021. An evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 110(3):563–572. doi: 10.1002/cpt.2350.
- Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE. 2012. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 92(4):414–417. doi: 10.1038/clpt.2012.96.
- WHO. 2023. Mental disorders. WHO Web site; [updated 08 June 2022; accessed 2023 April 20]. https://www.who.int/news-room/fact-sheets/detail/mental-disorders.

