ABSTRACT
Plants have evolved various belowground traits to adapt to the changing environments, and root-associated soil microbes play a crucial role in the response, adaptation, and resilience to adverse environmental conditions. This comprehensive review explores the diverse interactions between plants and soil microbes, focusing on the role of root-associated microbiota, with a particular emphasis on arbuscular mycorrhizal fungi, in plant responses to diverse environmental conditions. How plant genotype, root traits, and growth environments influence these interactions, and consequently plant resilience and productivity, are discussed. Recent advances in root phenotyping, including traditional and machine learning-based methods are also presented as an innovative tool to study and characterize root-microbe interactions. Overall, these studies highlight the importance of considering the hidden side of the interactions between roots and microbes to improve plant nutrition and protection in the context of sustainable agriculture in the face of climate change.
Highlights
Roots and associated microbes are relevant for improving plant nutrition and tolerance
AM symbiosis plays a role in plant responses to environmental stresses
Root phenotyping could represent an innovative tool to study root-microbe interactions
1. Introduction
Global climate change is predicted to alter environmental variables in an unprecedented manner, exacerbating soil degradation and affecting crop performance and productivity (Grayson Citation2013; Lynch et al. Citation2021). In several European countries, the simultaneous incidence of high average temperatures and water scarcity due to reduced rainfall are predicted to be the predominant adverse conditions according to the predictions (Moriondo et al. Citation2011). Together with drought, salinity and nutritional stresses are among the major threats that adversely affect plant growth and productivity (Lesk et al. Citation2016). Flood events also affect natural vegetation and crops, resulting in low O2 status in root tissues during waterlogging, but also in shoots when plants are completely submerged (Pedersen et al. Citation2017). To adapt to these challenges, plants have developed a variety of below-ground traits to access and efficiently uptake the available resources present in the soil (Freschet et al. Citation2018; Fontaine et al. Citation2023), and to mitigate the negative effects of abiotic factors such as drought, salinity, and flooding events on their growth and productivity. These below-ground traits encompass the structure and function of roots, which are defined as the parts of a plant that attaches it to the ground or to a support, typically underground, conveying water and nourishment to the rest of the plant via numerous branches and fibers (Oxford English Dictionary). Roots are, indeed, responsible for nutrient uptake and assimilation from the soil, although this process is not yet fully understood. Although several efforts have been devoted to develop several approaches for achieving more stress-tolerant and climate-flexible crops (González Guzmán et al. Citation2021), the valorization of a root trait, or a combination of traits, to optimize the resilience to environmental stresses has been overlooked. In addition to plant genotype, environmental factors such as nutrient and water availability, and the plant’s capacity to efficiently use soil resources, have a significant impact on plant growth under stress conditions. Improving crop productivity is directly dependent on an efficient root system, and identifying plant genotypes harboring root traits associated with better adaptations to a certain stress is one of the strategies being pursued by researchers and breeders (Lynch Citation2013). Architectural, anatomical, and physiological root traits are among the relevant traits for plant response and resilience to environmental changes (Nerva et al. Citation2022). Natural selection favors plants that drive root activity to optimally utilize the resources that are heterogenously distributed in soils. In addition, the ability of roots to interact with soil biota is also crucial for nutrient uptake from the soil (Galindo-Castañeda et al. Citation2022). Phenotypic plasticity is the ability of an organism to change its phenotype in response to diverse environmental conditions (Sultan Citation2000). It is worth noting that the plasticity of the root systems allows plants to adapt to a range of biotic and abiotic constraints that limit plant productivity (Schneider and Lynch Citation2020), and root-associated soil microbes play a fundamental role in adapting to adverse environmental conditions (Hou et al. Citation2021; Mesny et al. Citation2023). Different microbial functions, i.e. nutrient uptake, nitrogen cycle, and phosphorus solubilization, might act synergistically if properly associated with promising plant root genotypes (Galindo-Castañeda et al. Citation2022).
In this review, critical factors of the relationships among soil microbes, roots, and environmental conditions are introduced and discussed. These include the recruitment of microbes in stressed environments, a focus on the establishment and functioning of the arbuscular mycorrhizal (AM) symbiosis, particularly under abiotic stress conditions, and the significant role of root phenotyping techniques in advancing our understanding of plant-microbe interactions.
2. Microbe recruitment in stressed environments
It is now recognized that plant health can be conditioned by the diversity and structure of microbial communities associated with the holobiont (Bettenfeld et al. Citation2022). The composition and the diversity of microbial communities of the holobiont are different in plant organs (e.g. roots, shoots), but the soil is the main reservoir for the assemblage of plant organ microbiota (de Souza et al. Citation2016). Given the importance of soil nutrients for plant growth, we will focus on the root-associated microbiota present within, on the surface, and in the vicinity of the root (rhizosphere). The root exudates released by the roots vary considerably depending on the plant stage and species, but also among genotypes of the same species. They represent a source of carbon for the root-associated microbes, making the rhizosphere, i.e. the thin layer of soil surrounding plant roots, enriched in nutrients compared to the surrounding soil and more suitable for bacteria and fungi collectively quoted as the rhizosphere microbiota (Escudero-Martinez and Bulgarelli Citation2019). Indeed, it is well known that plant roots release a wide range of chemical compounds to recruit microbes from the rhizosphere (Huang et al. Citation2014). These root exudates moderate rhizospheric and multitrophic interactions at the species and at the community level (Sun et al. Citation2021; Wen et al. Citation2022). Root-associated microbes then influence plant health and growth through various mechanisms (Huang et al. Citation2014). Beneficial microbes living in the rhizosphere and in the root tissues (as endophytes) activate stress-protective pathways that help plants to alleviate the negative impact of environmental stresses, promoting several, and combined mechanisms that restrain the deleterious effects due to stress conditions. The chemistry in the rhizosphere is crucial for the selection of symbiotic associations. The metabolites released by the roots can play different roles: i.e. nutrients consumed by specific microbes, antimicrobial compounds produced against putatively pathogenic microbes, and signals to attract specific microbes (Thoms et al. Citation2021). Plants have the ability to actively modify the relative abundance of specific compounds in the exudates, thereby further acting on the composition of microbial communities in the rhizosphere (Zhalnina et al. Citation2018). Changes in microbial communities can impact plant phenology, influence the timing of plant flowering, and potentially increase reproductive fitness (Lu et al. Citation2018). The role of plant genotype on the interactions with soil microbial communities has been studied in various crops, such as barley (Bulgarelli et al. Citation2013, Citation2015), common bean (Pérez-Jaramillo et al. Citation2017, Citation2019), and maize (Schmidt et al. Citation2016). For example, higher recruitment of plant growth-promoting bacteria by ancient wheat varieties compared to modern and improved wheat varieties has been observed (Valente et al. Citation2020; Yue et al. Citation2023). The effects of plant domestication on the chemical diversity of the rhizosphere have been reported by Iannucci et al. (Citation2017). This work supported the hypothesis that root exudates, which are affected by the genotype and by the genotype-by-environment interaction such as the soil type, maintain and support a great specific microbial diversity in the rhizosphere associated with any type of plant species. Consistent with the genomic evidence for barley domestication shown by Bulgarelli et al. (Citation2015), data on root metabolites in wild emmer, domesticated emmer, and modern durum wheat suggested that selection during wheat domestication and modern breeding might have played a significant role in shifts of microbiome-plant interactions. Furthermore, Mahoney et al. (Citation2017) showed that different wheat cultivars differentially altered the bacterial abundances in the rhizosphere, and Essiane-Ondo et al. (Citation2019) showed that different wheat cultivars responded differently to native AM fungal communities, both in terms of diversity and yield. These observations, made in annual plants, have also been described in perennial plants such as grapevine (Noceto et al. Citation2024). This is consistent with the soil-dependent and genotype-dependent variations observed in the composition of rhizosphere microbial communities (Lareen et al. Citation2016). Indeed, microbiomes associated with wheat roots have changed through a breeding process (Kinnunen-Grubb et al. Citation2020), with Acidobacteria and Actinobacteria enriched in old cultivars, while taxa belonging to Candidatus Saccharibacteria, Verrucomicrobia, and Firmicutes enriched in modern ones. These last were also enriched in fungal pathogens, including Fusarium, Neoascochyta, and Microdochium genera (Kinnunen-Grubb et al. Citation2020). Focusing on maize (Zea mays), wheat (Triticum aestivum), and barley (Hordeum vulgare), Xiong et al. (Citation2021) clearly demonstrated that host selection shapes crop microbiome assembly and network complexity, having important implications for future crop management and manipulation of crop microbiome for sustainable agriculture. Bai et al. (Citation2013) assessed a diverse set of bread wheat germplasm and found that plant height was positively correlated to different evaluated root traits, such as total root length, seminal laterals length, seminal axes length, seminal laterals surface area, seminal laterals volume, and root dry weight. Additionally, environmental stresses can compromise the plant associations with beneficial microbes, thereby significantly limiting plant fitness (de Vries et al. Citation2020). It has been well explained that during a drought event, the interactions with soil-beneficial microbes, such as plant growth-promoting bacteria (PGPB) and AM fungi, improve drought tolerance. However, these relationships break down in the presence of severe or prolonged drought, and different plant-microbial associations are assembled after drought, with the potential to have an impact on future plant and soil drought responses (de Vries et al. Citation2020). Despite the importance of these microbial communities to plant growth, our understanding of the mechanisms driving microbiome assembly and composition remains incomplete, particularly in stressed environmental conditions and real-world scenarios. In tomatoes, for instance, it has been reported that two different genotypes showed contrasting responses to water deficit, primarily through diverse rhizosphere microbiota recruitment upon two diverse irrigation treatments (Sillo et al. Citation2022). Results suggested that, in open fields, bacterial communities in tomato roots were shaped by water deficit, leading to the selection of taxa potentially involved in promoting tolerance to drought, with differences between the two diverse genotypes (Sillo et al. Citation2022). Following a flood event, specific bacterial taxa were associated with a lentil landrace, as well as enhanced tolerance to fungal diseases, compared to commercial varieties (Brescia et al. Citation2023). Additionally, rootstock genotypes and an AM fungal inoculum have been shown to influence the microbes associated with grapevine roots, thus promoting plant growth and defense mechanisms (Nerva et al. Citation2022). Ecological aspects related to root exudates in drought conditions have also been studied in fast-growing and slow-growing plant species (i.e. the grass Holcus lanatus and the forb Rumex acetosa). The results showed that qualitative changes in root exudates increased the activity of the soil microbial community, supporting regrowth and recovery after drought (de Vries et al. Citation2019). Similarly, plants undergoing salinity stress recruit microbes from soil according to their ecological functions (Zheng et al. Citation2021).
Plants do not react to all the microbes living in the rhizosphere by activating defense responses, but they must distinguish between pathogens, mutualistics, and commensals, maintaining a balance between growth and defense (Schloter and Matyssek Citation2009; Thoms et al. Citation2021; ). Roots are continually exposed to the diverse microbes present in the surrounding soil, especially in the rhizosphere, and may undergo further selection (Philippot et al. Citation2013), investing energy in an immune or symbiotic response. Changes in root exudates under stress play a key role in the resistance to stress (Chai and Schachtman Citation2022), but also in the recruitment of specific soil microorganisms. When plants are exposed to stress, such as environmental challenges or nutrient deficiencies, they adapt by changing the composition of compounds released by their roots. The crosstalk between plants and microorganisms in the rhizosphere acts as signal molecules that select and recruit soil microorganisms to improve plant resource use efficiency under stress conditions (Rolfe et al. Citation2019).
Figure 1. The hidden root microbiome, a vast spectrum of microorganisms involved in interactions with roots and the rhizosphere.
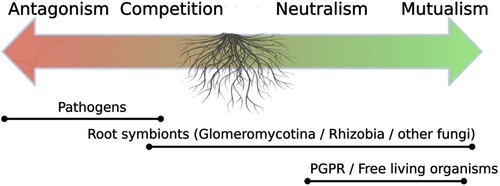
Among the molecules produced by plants during stress, strigolactones have been shown to play regulatory roles against abiotic stress in plants (Pandey et al. Citation2016; Ling et al. Citation2020). Strigolactones are a class of plant hormones involved in many processes of plant development and acclimation to environmental stress (Waters et al. Citation2017). In plant exudates, higher levels of strigolactones are associated with abiotic stresses and have been shown to affect the entire microbial community. Among the beneficial root-associated microbes, a relevant role is played by AM fungi, which are symbiotic soil-borne fungi that can impact plant development and health (Smith and Read Citation2008). Strigolactones have been shown to be involved in the crosstalk between the plant and AM fungi. Indeed, in mycorrhized tomato (Solanum lycopersicum) and lettuce (Lactuca sativa) plants, it has been shown that AM colonization and strigolactone production is stimulated under drought (Ruiz-Lozano et al. Citation2016). These results suggest a positive correlation between strigolactone production and drought tolerance in AM-colonized plants as a ‘call for help’ to promote the establishment of AM symbiosis (Ruiz-Lozano et al. Citation2016).
3. The arbuscular mycorrhizal symbiosis establishment and functioning
The arbuscular mycorrhizal symbiosis establishment
AM fungi recognize host plants through the perception of root-secreted strigolactones that promote spore germination, hyphal growth and branching, mitochondrial activity in the pre-symbiotic mycelium, the expression of effector genes, and the release of plant-directed chemical signals known as Myc-factors. Currently characterized Myc-factors include chitin-derived molecules, such as chitooligosaccharides and lipo-chitooligosaccharides, which act as microbe-associated molecular patterns (MAMPs). It is worth noting that recently lipo-chitooligosaccharides were reported to be produced by a wide range of fungi belonging to divergent lineages, where they might act as signals in growth and development (Rush et al. Citation2020), in addition to have a role in plant recognition. The perception of Myc-factors by plant receptors induces the activation of the common SYMbiosis pathway (SYM pathway), which triggers the expression of specific genes of the symbiosis and induces a massive reorganization of cell structure (Pimprikar and Gutjahr Citation2018; Volpe et al. Citation2023). After the molecular cross-talk, the activation of the SYM pathway leads to the expression of the transcription factor LjRAM1, resulting in the expression of specific genes involved in the development of highly branched arbuscules and in nutrient transport (e.g., phosphate and nitrogen transporters, plant carbohydrate and lipid metabolism) (Garcia et al. Citation2016; Pimprikar et al. Citation2016; Rich et al. Citation2017). Furthermore, colonization with AM fungi induces the upregulation of defense genes, suggesting that defense proteins are involved in mycorrhizal infection, but also could confer higher resistance to root pathogens (Goddard et al. Citation2021). Recently, Ca2+-mediated signaling mechanisms discriminating plant immunity- and symbiosis-related pathways have been deciphered (Binci et al. Citation2023).
The trophic base of arbuscular mycorrhiza
Improving the mineral nutrition of the plant and the carbon nutrition of AM fungi are considered to be the main benefits of AM symbiosis. This applies in particular to improving the phosphorus and nitrogen nutrition of AM plants, as these two macroelements are essential to plants in large quantities and are often present in limited available concentrations in natural environments (Elser et al. Citation2007). Plants compatible with the AM symbiosis are supposed to have, in addition to the direct uptake of nutrients by the epidermal cells of the roots, specific ‘mycorrhizal uptake pathways’ to absorb the mineral elements taken up from the soil by the AM fungal partner. An extensive review of the mycorrhizal pathways and transport systems involved in nutrient exchange has recently been published (Casieri et al. Citation2013; Wipf et al. Citation2019).
AM fungi form common mycorrhizal networks (CMNs), capable of affecting plant behavior, development, and physiology (Wipf et al. Citation2019). A CMN is established when the roots of several plants are connected by an underground network formed by the mycelium of AM fungi (Kiers et al. Citation2011). CMNs can be exclusive when roots of the same plant species form mycorrhizae with the same AM fungi, or inclusive when roots of different plant species are connected by the same AM fungi due to their low host specificity (Wipf et al. Citation2019). Physical evidence for the presence of CMN among plants was established nearly half a century ago (Reid and Woods Citation1969). Evidence on the role and place of CMN in ecosystem functioning has only really been studied in the last three decades, but not extensively (Simard et al. Citation1997; Fitter et al. Citation1998). Links between plants via CMN have been reported in studies showing the transfer of isotopes (13C,33P, and 15N) from one plant to another by autoradiography (Francis and Read Citation1984; Newman et al. Citation1994; Walder et al. Citation2015; Calabrese et al. Citation2019). CMN should not be thought of as straight motorways connecting two plants, but as a real motorway network where roads cross and join. Fungal hyphae, and AM fungi in particular, can fuse, forming anastomoses (physical connection of hyphae leading to the pooling of cytoplasms and their contents) between isolated filaments and existing CMN when these are genetically close (Giovannetti et al. Citation2001). These connections, or anastomoses, increase the plant’s associated functions (Mikkelsen et al. Citation2008).
4. Arbuscular mycorrhizal symbiosis and environmental stresses
The growing population and rising food demand in the context of climate change, soil depletion and competition among different land uses, and the requirement to use less chemical fertilizers and pesticides are only a few of the issues that modern agriculture must overcome (Gupta et al. Citation2020). Although the response to salinity mediated by AM fungi has been evaluated in diverse host plants (Pollastri et al. Citation2018; Duc et al. Citation2021; Ma et al. Citation2022), most of the published studies focused on the impact of the AM symbiosis on plant response to water deficit in different crops (Dell’Amico et al. Citation2002; Subramanian et al. Citation2006; Aroca et al. Citation2008; Wang et al. Citation2015; Symanczik et al. Citation2015; Zhang et al. Citation2015; Chitarra et al. Citation2016; Ruiz-Lozano et al. Citation2016; Recchia et al. Citation2018; Rivero et al. Citation2018; Volpe et al. Citation2018). However, the pathways involved in the improvement of the water deficit tolerance by AM fungi are complex. Multiple plant responses are involved, along with the responses induced by the stress condition in the AM fungus (Cheng et al. Citation2021; Shi et al. Citation2023). Plant-beneficial symbionts can be in fact directly affected by the stresses itself, or indirectly by plant stress responses (Bastías et al. Citation2023). Due to the complexity of the response to stress, which involves both partners, the mechanisms underlying the enhanced tolerance mediated by the AM symbiosis are still to be fully discovered. Several positive impacts have been reported on photosynthetic traits in host plants, showing improved performances upon different environmental stresses such as drought, salinity, and extreme temperature (Balestrini et al. Citation2020). Differences between tomato responses in plants inoculated with two AM fungal species have been reported (Chitarra et al. Citation2016; Volpe et al. Citation2018), suggesting that the response could be genotype-dependent. Particularly, Rhizophagus intraradices led to an improved intrinsic water use efficiency (iWUE) with respect to Funneliformis mosseae and control uninoculated plants (Symanczik et al. Citation2020). It has been reported that AM fungal symbiosis improves plant water content by the mycorrhizal root system through extraradical mycelium: AM hyphae can reach water resources from the soil both by exploring micropores not accessible to host roots and by affecting soil structure (Ruiz-Lozano and Azcón Citation1995; Augé et al. Citation2008). The plant physiology improvement by the AM symbiosis upon water-limiting conditions has been extensively reported in diverse crops (Ruiz-Lozano et al. Citation2015; Augé et al. Citation2015; Balestrini et al. Citation2020), including an increase in hydraulic conductivity (Quiroga et al. Citation2019). At the root level, the presence of the AM fungus leads to extensive modifications in root physiology, as well as in root cells to accommodate the fungal structures, and these changes are controlled by specific gene expression patterns in the host plant (Balestrini and Bonfante Citation2005; Fiorilli et al. Citation2009; Guether et al. Citation2009; Vangelisti et al. Citation2018; Balestrini et al. Citation2019). During water deficit it has been demonstrated, through targeted gene expression approaches, that diverse drought-responsive genes are differentially regulated in AM-colonized tomato and sorghum plants (both in roots and leaves) (Chitarra et al. Citation2016; Ruiz-Lozano et al. Citation2016; Symanczik et al. Citation2020) as well as genes involved in nutritional aspects (Symanczik et al. Citation2018; Volpe et al. Citation2018; Fiorilli et al. Citation2022). Among the genes that are differentially regulated in mycorrhizal roots, several researches focused attention on the expression of plant genes coding for aquaporins (AQPs). The AM symbiosis-mediated regulation is complex and is dependent on the plant and fungal species involved in the symbiosis and on the level and duration of the stress (Symanczik et al. Citation2020). Additionally, the simultaneous induction of both fungal and plant AQP genes in roots confirms that the two symbionts strictly cooperate to regulate the drought-stress response (Chitarra et al. Citation2016). Looking at nutritional aspects of the symbiosis upon water deficit conditions, Volpe et al. (Citation2018) focused the attention on tomato and fungus phosphate transporter (PT) genes, showing a different regulation of the considered PT genes by two AM fungi. Fiorilli et al. (Citation2022) have recently demonstrated that AM fungi positively affected sulfur homeostasis under water deficit in durum wheat but with differences between the two considered cultivars (Svevo and Etrusco). Mycorrhizal symbiosis positively impacted water status and nutrient uptake in Svevo compared to Etrusco cultivars (Fiorilli et al. Citation2022). The results of this work underscore the importance of considering nutrient use efficiency and the influence of other biotic soil components, such as AM fungi, when identifying drought-tolerant plants. This suggests that focusing exclusively on drought tolerance without considering these factors may not provide a comprehensive assessment of plant resilience (Fiorilli et al. Citation2022; Nerva et al. Citation2022). Diverse -omics approaches, such as transcriptomics (Balestrini et al. Citation2019; Keller-Pearson et al. Citation2023) and metabolomics (Rivero et al. Citation2018), have been also used to highlight the responses to drought response-mediated by AM symbiosis. It is worth noting that the AM symbioses led to an increase in plant metabolic plasticity to cope with stress, with diverse responses that were common to the different tested AM fungi, while others were exclusively linked to particular isolates. A relevant metabolism reprogramming was mainly observed in salt stress conditions, mirroring the improved tolerance to stress (Rivero et al. Citation2018). The AM symbioses enhanced plant metabolic plasticity in response to stress, eliciting both common responses across different tested AM fungi and exclusive responses specific to particular isolates. A significant metabolic remodeling was observed under salt stress conditions, reflecting an enhanced tolerance to such stress (Rivero et al. Citation2018).
Although some aspects of the relationship between symbiosis and response to environmental stress have been elucidated, the involved mechanisms are complex and need to be further explored. Keller-Pearson et al. (Citation2023) for example have recently reported that carrot roots and R. irregularis show a diverse regulation of gene expression during drought, with carrot reducing its apparent investment in symbiosis and the fungus increasing its apparent symbiotic efforts. However, cooperation between host plants and AM fungi is often dependent on the symbiotic partners, and it is related to several factors, such as environmental conditions, resources availability, and the functional diversity of the symbiotic partners. Looking at the fungal side, a highly conserved transcription factor in Rhizophagus irregularis, namely RiMsn2, has recently been reported to play a crucial role in the fungal response to drought (Fan et al. Citation2023). Experiments in transformed yeasts showed that this gene is key to maintain osmotic balance and regulate trehalose content, and Virus-induced gene silencing (VIGS) experiments in R. irregularis revealed that it is also crucial for the formation of arbuscules (Fan et al. Citation2023).
It is worth noting that variation in the results is explained by a diverse response to AM symbiosis and contrasting results in terms of perceived benefits (i.e. biomass and nutrient acquisition, stress tolerance) when different plant or fungal genotypes are considered. Diverse combinations can in fact have different colonization rates as well as a diverse response to stress.
Response to different combinations of arbuscular mycorrhizal fungi during a stress
It is well known that the benefits provided by AM symbiosis depend on the AM fungal species involved as partners (van der Heijden et al. Citation1998; Powell and Rillig Citation2018). Specific benefits provided by AM fungal species have been attributed to certain functional traits such as hyphal exploration distance, hyphal uptake capacity and strategy, and regenerative strategies (Chagnon et al. Citation2013; Chen et al. Citation2018). In a meta-analysis collecting data from more than 3000 studies of non-mycorrhizal plants versus mycorrhizal plants inoculated with a single AM fungal species under different abiotic stresses, Marro et al. (Citation2022) have shown that various taxonomic groups may provide different nutritional benefits depending on the type of stress. Previously, these traits were shown to be phylogenetically conserved, but differ among three main taxonomic families (Gigasporaceae, Glomeraceae, and Acaulosporaceae) (Hart and Reader Citation2002; De La Providencia et al. Citation2005; Powell et al. Citation2009; Koch et al. Citation2017). However, the taxonomic group does not explain the entire diversity of functional traits observed at the species and strain level. Indeed, the ability of plants to respond to AM fungi in terms of performance depends on the plant-fungus genotype combination, and the abiotic and biotic environment (Berger and Gutjahr Citation2021). Recently, the transcriptional and physiological responses of sorghum to two different AM fungal species, R. irregularis and F. mosseae, under 16 different conditions of nitrogen (N) and phosphorus (P) supply has been investigated. Physiological and gene expression patterns of ammonium and phosphate transporters reveal fine-scale differences between two AM fungal species in the nutritional interactions with sorghum plants. Evidence showed that AM fungal species differ in their strategy for nutrient transfer to the plant, suggesting that different species may harbor different ecological roles (Boussageon et al. Citation2022). In addition, different AM fungi exhibit different capacities to elevate the tolerance to salinity and drought. In Casuarina plants, three strains of Rhizophagus (R. irregularis DAOM197198, R. aggregatus DAOM2277128, and R. fasciculatus) have been tested under salt stress conditions with R. fasciculatus the most performant (Djighaly et al. Citation2018). In sorghum, R. irregularis DAOM197198 and R. arabicus were tested for their capacity to increase drought tolerance. R. arabicus, a strain endemic to hyper-arid ecosystems, was shown to significantly increase sorghum nutrition during drought stress (Symanczik et al. Citation2018). This study highlights the importance of selecting strains adapted to a given environment for better plant response to stress.
In a metanalysis, Rúa et al. (Citation2016) showed that sympatric relationships among plants, AM fungi, and soil tend to outperform allopatric relationships. If these three components coexist in the same geographical region, the interactions are often more advantageous, resulting in better absorption of nutrients. These results suggest that the local adaption of the plant and the community of AM fungi to a novel environment should be considered while designing experimentation under drought and salinity stresses.
5. Interactions between arbuscular mycorrhizal fungi and other beneficial microbes of the root microbiota
Recent studies have shown that services provided by AM fungi (Gianinazzi et al. Citation2010) are often facilitated by the microbiota in the soil or associated with the AM fungi or the plant. Multitrophic interactions among roots, AM fungi and certain soil bacteria are established, the beneficial effects of which may be synergistic and additive (). Some evidences have also shown the suppression of some AM fungal activities by soil microbiota (Svenningsen et al. Citation2018). Besides AM fungi, other microbes as plant growth-promoting rhizobacteria (PGPRs) can also have an impact on plant development and health. PGPRs can be either free-living, root-bound, bound to fungal hyphae, or bound to spores (Bianciotto et al. Citation1996). PGPRs adhere to roots or AM fungi via biofilms formed from glycoproteins. PGPRs are bacteria belonging to various genera, including Pseudomonas, Bacillus, Rhizobia, and Azotobacter spp. (Benizri et al. Citation2001). Some of them act directly on AM fungi, either in vitro (Ordoñez et al. Citation2016) or in soil (Battini et al. Citation2017), because they (i) improve root colonization (Pivato et al. Citation2009), (ii) promote hyphal growth (Bidondo et al. Citation2011); (iii) facilitate or improve spore germination by degrading chitin, the main component of AM fungal spore walls (Roesti et al. Citation2005).
Figure 2. Conceptual diagram of the hidden soil interactome and its consequences for plant nutrition (
 ). A: plant alone, B: plant + Plant Growth Promoting Rhizobacteria, C: plant + Arbuscular Mycorrhizal Fungi, D: plant + Plant Growth Promoting Rhizobacteria + Arbuscular Mycorrhizal Fungi.
). A: plant alone, B: plant + Plant Growth Promoting Rhizobacteria, C: plant + Arbuscular Mycorrhizal Fungi, D: plant + Plant Growth Promoting Rhizobacteria + Arbuscular Mycorrhizal Fungi.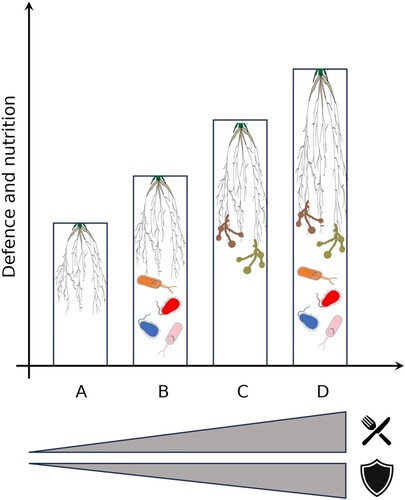
The taxonomic diversity of PGPRs in the different ‘spheres’ mentioned above is extremely variable; this diversity depends in particular, as mentioned previously, on the composition of root exudates, but also on the composition of exudates from the hyphae of AM fungi.
PGPRs stimulate mycorrhization when growth is constrained or inhibited by extreme soil and climatic conditions such as drought (Vivas et al. Citation2005). PGPRs stimulate root colonization by AM fungi by repressing phytopathogen-related signaling pathways (Gamalero et al. Citation2008; Stearns et al. Citation2012). Finally, they stimulate root colonization by AM fungi by promoting the release of plant flavonoids that direct hyphal growth (Xie et al. Citation1995). If the presence of PGPRs is beneficial for AM fungi, so is the reciprocal; Gamalero et al. (Citation2004) showed that F. mosseae improved the long-term survival of a strain of P. fluorescens.
Although there is little data on the combined role of PGPRs and AM fungi on plant response (for example see Vosatka et al. Citation1992; Gamalero et al. Citation2004; Ordookhani and Zare Citation2011; Dohroo and Sharma Citation2012; Tavasolee et al. Citation2013; Berta et al. Citation2014; Bona et al. Citation2014), Todeschini et al. (Citation2018) recently reported the impact on strawberry production and quality of PGPR and AM fungal co-inoculation as well as the importance of the used bacterial and fungal strains. The effects of interactions between roots, AM fungi and PGPRs can be exploited for the benefit of sustainable agriculture (Noceto et al. Citation2021). Since AM fungi and PGPRs are beneficial microorganisms, their synergistic or additive effects could be even more beneficial in improving yield and quality, as well as controlling certain diseases.
The question of the extent of the impact of the rhizosphere is still under debate, especially when considering the plant holobiont and the extra-radical mycelium of AM fungi as an extension of the root system, leading to the formation of the hyphosphere specifically. The hyphosphere microbiome has diverse functions and the recruited microbial species could be different (Wang et al. Citation2022). Regarding plant phosphate nutrition and the weak ability of AM fungi to solubilize mineral forms of inorganic phosphate alone (Antunes et al. Citation2007), AM fungi recruit other microbes with phosphate degradation capabilities (Zhang et al. Citation2022).
6. Approaches of root phenotyping for plant interactions
The importance of root phenotyping is growing as it plays a pivotal role in understanding how plants respond to different environmental conditions and stress factors (Amtmann et al. Citation2022). Root phenotyping approach to examine plant responses to various abiotic and biotic conditions, including interactions with soil microbes, holds great promise for advancing our understanding of crop resilience (). Nowadays, root phenotyping in controlled conditions can be conducted in both two- and three-dimensions (2D and 3D). 2D root phenotyping typically involves the analysis of images or scans of roots on a flat surface, while 3D root phenotyping involves capturing and analyzing three-dimensional images or models of root structures (Atkinson et al. Citation2019). Traditional nondestructive techniques for studying root growth, such as rhizotrons, typically produce 2D images (Atkinson et al. Citation2019). In addition to rhizotrons, one of the main tools to assess root phenotypes is represented by RhizoTubes (Jeudy et al. Citation2016). These tubes are innovative cylindrical rhizotrons designed for dynamic root phenotyping. They consist of concentric tubes that create separate zones for root growth, substrate, and nutrient solution, with a permeable membrane allowing the exchange of nutrients, water, as well as soil microbes, while preventing root passage (Jeudy et al. Citation2016). This design confines the root system in two dimensions, facilitating high-quality imaging using a dedicated camera (namely ‘RhizoCab’). RhizoTubes have proven to be useful also for studying interactions between roots and microbes (Jeudy et al. Citation2016). The high-resolution imaging capabilities of RhizoTubes might allow the assessment of root nodules (symbiosis with rhizobia) as well as the visualization of mycelium in the case of fungi interacting with roots (Jeudy et al. Citation2016). RhizoTubes might be used to perform noninvasive measurements of root development upon the impact of microbial strains/isolates, populations, or communities, as they could allow the inoculation of both symbiotic and free-living microbes on the substrate (Jeudy et al. Citation2016). The effect on roots of the bacterial strain Pseudomonas fluorescens C7R12 inoculated on pea and wheat roots was successfully studied in RhizoTubes (Jeudy et al. Citation2016). Another example is represented by the integrated system known as GrowScreen-PaGe that was employed to investigate the growth patterns of the model cereal Brachypodium distachyon Bd21-3 in the presence of the PGPR Azospirillum brasilense Sp245 (Gioia et al. Citation2016; Schillaci et al. Citation2021). This platform allowed the high-throughput phenotyping of both shoot and root growth under different abiotic conditions. Dedicated softwares for assessing nodulation in roots have also been developed. For example, the standardized quantitative analysis of nodulation patterns in legumes developed by Remmler et al. (Citation2014) allows for a standardized representation of the nodulated root system and calculates nodulation degree and root variables (Remmler et al. Citation2014). Additionally, a Soybean Nodule Acquisition Pipeline (SNAP) for nodule quantification based on deep learning detection and segmentation, in order to phenotype nodules on soybean roots at a higher throughput has been recently developed (Jubery et al. Citation2021). This pipeline was successfully used to assess the interactions between soybean and Bradyrhizobium bacteria (Jubery et al. Citation2021). As for the innovations in root growth analysis, advancements have also been made in the quantification of root colonization by AM fungi in root samples. Conventional methods for quantifying root colonization by AM fungi and hyphal abundance involve root samples staining of root fragments with lactic blue or other dyes for the labeling of the intraradical fungal structures, microscopic observations of the AM fungal-specific structures, and human scoring, which can be subjective and could vary between observers (Vierheilig et al. Citation2005; Sportes et al. Citation2022). The Trouvelot method is the traditional method for quantifying AM fungal colonization in roots, and it involves assessing root segments in glass slides through microscopical observations of the colonization percentage (Trouvelot et al. Citation1986). Another established method, i.e. the grid-line intersect method by Giovannetti and Mosse (Citation1980), is widely used to evaluate the rate of AM fungal root colonization by observing the presence of fungal hyphae. The use of root phenotyping systems, such as WinRHIZO™, has proven to be a promising method for obtaining accurate quantification of AM fungal structures in roots. WinRHlZO™ system (Regent Instruments Inc., Quebec, QC; Arsenault et al. Citation1995) and EZ-Rhizo software (Armengaud et al. Citation2009) are two of the most popular tools to analyze data from 2D root phenotyping. WinRHlZO™ utilizes a scanner to capture images and analyze global root measurements, such as average diameter, total length, and area, along with topology analyses of root system architecture (Arsenault et al. Citation1995). On the other hand, the EZ-Rhizo software is a semi-automated tool for measuring various aspects of lateral roots, including length, angle, and number (Armengaud et al. Citation2009) as well as the more recent free and open-source software RhizoVision Explorer (Seethepalli et al. Citation2021). The WinRHIZO™ has been proposed as a tool to estimate AM fungal root colonization and was validated with AM roots of Chengiopanax sciadophylloides when compared to the grid-line intersect method (R2 = 0.94; Deguchi et al. Citation2017). This method allows analyzing a large number of samples and facilitates comparison of results across different researchers by sharing analytical settings via a cac formatted file, i.e. the standard file generated by the software (Deguchi et al. 2017). The WinRHIZO™ method efficiently assessed AM fungal structures automatically, showing promise for further development and widespread use in evaluating AM fungi across different plant species (Deguchi et al. 2017). Recent advancements in machine learning, coupled with the availability of specialized softwares for root analysis, have promoted the emergence of innovative and less time-consuming methodologies for assessing root colonization by AM fungi. Recently, the Automatic Mycorrhiza Finder (AMFinder) software was developed as an innovative tool employing computer vision and convolutional neural networks for the automatic identification and quantification of AM fungal root colonization and intraradical hyphal structures in ink-stained root images (Evangelisti et al. Citation2021). This tool demonstrated high efficiency in providing high-confidence predictions across different datasets, including results on root samples of Nicotiana benthamiana, Medicago truncatula, Lotus japonicus, and Oryza sativa. A protocol was established for sample preparation and imaging, allowing the quantification of several AM fungal species through scanning or digital microscopy, including the observation of dynamic increases in colonization across the whole root systems over time (Evangelisti et al. Citation2021). Other approaches based on machine learning, i.e. machine learning-based procedure of the Zeiss Zen Intellesis software (Carl Zeiss Microscopy GmbH Jena, DE), were also evaluated, showing high efficiency and a high degree of correlation compared to traditional methods (Schillaci et al. Citation2020). More recently, Muta et al. (Citation2023) introduced a novel web-based application called ‘Tool for Analyzing root images to calculate the Infection rate of arbuscular Mycorrhizal fungi’ – TAIM, which employs machine learning classifiers to automatically determine the degree of AM fungal root colonization from 40x magnification microscopic images.
Table 1. Root phenotyping tools with a potential for studying root-microbe interactions. AMF: AM fungi.
It should be noted that AM fungal colonization of plant roots is a stepwise process influenced by environmental factors as well as by plant and AM fungal identity (Montero et al. Citation2019). Since the AM fungal colonization process is dynamic, with the presence of several colonization stages in the same root fragment, a spatial–temporal resolution approach, including multiple time points, should be employed to capture the development of specific AM fungal structures (Montero et al. Citation2019). Hence, in addition to microscopic observations for AM fungal colonization assessment, the monitoring of transcript levels of plant and AM fungal marker genes induced at different symbiotic stages should be used as a complementary analysis (Montero et al. Citation2019). While the use of AM fungal housekeeping genes as an indirect method to assess fungal biomass is common, the assessment of the expression of specific AM fungal marker genes regulated at distinct symbiotic stages is recommended (Balestrini et al. Citation2007; Helber et al. Citation2011; Tsuzuki et al. Citation2016; Sun et al. Citation2018). On the host plant side, time-course transcriptomic studies have shown genes associated with the early and late stages of symbiotic interaction (Choi et al. Citation2018; Pimprikar and Gutjahr Citation2018). Thus, it has been proposed that root samples should be collected at least two different times to obtain the initial and final stages of AM fungal colonization (Montero et al. Citation2019). The assessment of AM fungal colonization in roots can be also accomplished by using DNA-based molecular techniques on colonized roots. Quantitative polymerase chain reaction (qPCR) offers an efficient high-throughput tool, by using general AM fungal primers and either relative or absolute quantification methods, to assess AM fungal colonization in roots (Gollotte et al. Citation2004; Thonar et al. Citation2012; Hewins et al. Citation2015; Badri et al. Citation2016; Voříšková et al. Citation2017). Recently, a study by Corona Ramírez et al. (Citation2023) validated the use of a primer set (AMG1F and AM1) for qPCR in Petunia plants and tested on wheat, tomato, and leek plants. This tool proved to be useful in AM fungal colonization assessment and also highlighted that primer efficacy may vary with different crops and must be validated for each single tested plant species (Corona Ramírez et al. Citation2023). Additionally, it is worth noting that novel approaches to track the progression of AM symbiosis have been developed, such as pigment-based marker systems, as a useful alternative to conventional methods (Timoneda et al. Citation2021; Kumar et al. Citation2022) that rely on the generation of transgenic plants expressing mycorrhizal specific reporters, such as MycoRed, a betalain-based reporter system (Timoneda et al. Citation2021), and a method based on anthocyanin pigmentation (Kumar et al. Citation2022). It is worth noting that nondestructive three-dimensional (3D) phenotyping of roots in soil can also be currently accomplished by using tomographic techniques. These methodologies comprise X-ray computed tomography (X-ray CT) and magnetic resonance imaging (MRI) (Pfeifer et al. Citation2015; Pflugfelder et al. Citation2017). Both X-ray CT and MRI can be used to monitor root system growth over time without affecting plant development (Atkinson et al. Citation2019). However, it has been reported that potential negative effects of repeated X-ray CT scans on root development and soil biota may occur (Zappala et al. Citation2013).
Phenotyping studies of root-microbe interactions in the field are still deemed as a challenging task. However, high-resolution Minirhizotrons have been successfully used to explore root-fungal dynamics, focusing on the abundance and growth of plant fine roots and fungal mycelium (Defrenne et al. Citation2020). Results showed that in warmer, drier peat, ericaceous shrub roots, and ectomycorrhizal fungal rhizomorphs were more abundant, while in colder, wetter peat, fine roots of trees, ectomycorrhizal fungi, and dark-colored fungal hyphae were more abundant (Defrenne et al. Citation2020). By employing a Minirhizotron system, the peanut root systems, including nodule development, over time, have been analyzed (Rowland et al. Citation2015). Minirhizotrons indeed represented one of the most widely used and effective noninvasive tools for evaluating root growth under field conditions to date (Taylor Citation1987). Modern Minirhizotron systems typically include transparent tubes in the soil, a miniature video camera for capturing images, and a computer to control the camera and store images (Johnson and Meyer Citation1998). Future improvements in both hardware (imaging capturing cameras) and software (for example, by using a machine-learning image analysis pipeline; Bauer et al. Citation2022) will promote the study of roots interacting with soil microbes directly in situ.
Conclusion
In recent years, researchers have aimed to engineer the positive interaction between AM fungi and other soil microorganisms by co-inoculation to stimulate plant nutrition and plant protection. The critical point in plant development is the balance between plant nutrition and plant protection, and to which extent root microbiome could participate in such functions. The question of the symbiotic traits that need to be quantified is crucial. These traits need to be identified separately in microbial species and to be stable when the microbe is present in a microbial community.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Amtmann A, Bennett MJ, Henry A. 2022. Root phenotypes for the future. Plant Cell Environ 45:595–601. doi:10.1111/pce.14269.
- Antunes PM, Schneider K, Hillis D, Klironomos JN. 2007. Can the arbuscular mycorrhizal fungus Glomus intraradices actively mobilize P from rock phosphates? Pedobiologia. 51:281–286. doi:10.1016/j.pedobi.2007.04.007.
- Armengaud P, Zambaux K, Hills A, Sulpice R, Pattison RJ, Blatt MR, Amtmann A. 2009. EZ-rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J 57:945–956. doi:10.1111/j.1365-313x.2008.03739.x.
- Aroca R, Vernieri P, Ruiz-Lozano JM. 2008. Mycorrhizal and non-mycorrhizal Lactuca sativa plants exhibit contrasting responses to exogenous ABA during drought stress and recovery. J Exp Bot 59:2029–2041. doi:10.1093/jxb/ern057.
- Arsenault J-L, Poulcur S, Messier C, Guay R. 1995. WinrhlzoTM, a root-measuring system with a unique overlap correction method. Hort. Sci 30:906. doi:10.21273/hortsci.30.4.906d.
- Atkinson JA, Pound MP, Bennett MJ, Wells DM. 2019. Uncovering the hidden half of plants using new advances in root phenotyping. Curr Opin Biotechnol 55:1–8. doi:10.1016/j.copbio.2018.06.002.
- Augé RM, Toler HD, Sams CE, Nasim G. 2008. Hydraulic conductance and water potential gradients in squash leaves showing mycorrhiza-induced increases in stomatal conductance. Mycorrhiza. 18:115–121. doi:10.1007/s00572-008-0162-9.
- Augé RM, Toler HD, Saxton AM. 2014. Arbuscular mycorrhizal symbiosis alters stomatal conductance of host plants more under drought than under amply watered conditions: a meta-analysis. Mycorrhiza. 25:13–24. doi:10.1007/s00572-014-0585-4.
- Badri A, Stefani FOP, Lachance G, Roy-Arcand L, Beaudet D, Vialle A, Hijri M. 2016. Molecular diagnostic toolkit for Rhizophagus irregularis isolate DAOM-197198 using quantitative PCR assay targeting the mitochondrial genome. Mycorrhiza. 26:721–733. doi:10.1007/s00572-016-0708-1.
- Bai C, Liang Y, Hawkesford MJ. 2013. Identification of QTLs associated with seedling root traits and their correlation with plant height in wheat. J Exp Bot 64:1745–1753. doi:10.1093/jxb/ert041.
- Balestrini R, Bonfante P. 2005. The interface compartment in arbuscular mycorrhizae: a special type of plant cell wall? Plant Biosyst 139:8–15. doi:10.1080/11263500500056799.
- Balestrini R, Brunetti C, Chitarra W, Nerva L. 2020. Photosynthetic traits and nitrogen uptake in crops: which is the role of arbuscular mycorrhizal fungi? Plants. 9:1105. doi:10.3390/plants9091105.
- Balestrini R, Gómez-Ariza J, Lanfranco L, Bonfante P. 2007. Laser microdissection reveals that transcripts for five plant and one fungal phosphate transporter genes are contemporaneously present in arbusculated cells. MPMI. 20:1055–1062. doi:10.1094/mpmi-20-9-1055.
- Balestrini R, Rosso LC, Veronico P, Melillo MT, De Luca F, Fanelli E, Colagiero M, di Fossalunga AS, Ciancio A, Pentimone I. 2019. Transcriptomic responses to water deficit and nematode infection in mycorrhizal tomato roots. Front Microbiol 10:1807. doi:10.3389/fmicb.2019.01807.
- Bastías DA, Johnson LJ, Applegate ER, Jáuregui R, Card SD. 2023. Tripartite associations: a bacterial symbiont of fungi promotes plant growth without compromising the benefits conferred by an Epichloë endophyte. Envir. Exp. Bot. 215:105510. doi:10.1016/j.envexpbot.2023.105510.
- Battini F, Grønlund M, Agnolucci M, Giovannetti M, Jakobsen I. 2017. Facilitation of phosphorus uptake in maize plants by mycorrhizosphere bacteria. Sci Rep 7. doi:10.1038/s41598-017-04959-0.
- Bauer FM, Lärm L, Morandage S, Lobet G, Vanderborght J, Vereecken H, Schnepf A. 2022. Development and validation of a deep learning based automated minirhizotron image analysis pipeline. Plant Phenomics. 2022:9758532. doi:10.34133/2022/9758532.
- Benizri E, Baudoin E, Guckert A. 2001. Root colonization by inoculated plant growth-promoting rhizobacteria. Biocontrol Sci Technol 11:557–574. doi:10.1080/09583150120076120.
- Berger F, Gutjahr C. 2021. Factors affecting plant responsiveness to arbuscular mycorrhiza. Curr Opin Plant Biol 59:101994. doi:10.1016/j.pbi.2020.101994.
- Berta G, Copetta A, Gamalero E, Bona E, Cesaro P, Scarafoni A, D’Agostino G. 2013. Maize development and grain quality are differentially affected by mycorrhizal fungi and a growth-promoting pseudomonad in the field. Mycorrhiza 24:161–170. doi:10.1007/s00572-013-0523-x.
- Bettenfeld P, Cadena i Canals J, Jacquens L, Fernandez O, Fontaine F, van Schaik E, Courty P-E, Trouvelot S. 2022. The microbiota of the grapevine holobiont: a key component of plant health. J Adv Res 40:1–15. doi:10.1016/j.jare.2021.12.008.
- Bianciotto V, Bandi C, Minerdi D, Sironi M, Tichy HV, Bonfante P. 1996. An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. Appl Environ Microbiol 62:3005–3010. doi:10.1128/aem.62.8.3005-3010.1996.
- Bidondo LF, Silvani V, Colombo R, Pérgola M, Bompadre J, Godeas A. 2011. Pre-symbiotic and symbiotic interactions between Glomus intraradices and two Paenibacillus species isolated from AM propagules. In vitro and in vivo assays with soybean (AG043RG) as plant host. Soil Biol Biochem 43:1866–1872. doi:10.1016/j.soilbio.2011.05.004.
- Binci F, Offer E, Crosino A, Sciascia I, Kleine-Vehn J, Genre A, Giovannetti M, Navazio L. 2023. Spatially and temporally distinct Ca2 + changes in Lotus japonicus roots orient fungal-triggered signalling pathways towards symbiosis or immunity. J Exp Bot 75:605–619. doi:10.1093/jxb/erad360.
- Bona E, Lingua G, Manassero P, Cantamessa S, Marsano F, Todeschini V, Copetta A, D’Agostino G, Massa N, Avidano L, et al. 2014. AM fungi and PGP pseudomonads increase flowering, fruit production, and vitamin content in strawberry grown at low nitrogen and phosphorus levels. Mycorrhiza 25:181–193. doi:10.1007/s00572-014-0599-y.
- Boussageon R, Marro N, Janoušková M, Brulée D, Wipf D, Courty P-E. 2022. The fine-tuning of mycorrhizal pathway in sorghum depends on both nitrogen−phosphorus availability and the identity of the fungal partner. Plant Cell Environ 45:3354–3366. doi:10.1111/pce.14426.
- Brescia F, Sillo F, Franchi E, Pietrini I, Montesano V, Marino G, Haworth M, Zampieri E, Fusini D, Schillaci M, et al. 2023. The ‘microbiome counterattack’: insights on the soil and root-associated microbiome in diverse chickpea and lentil genotypes after an erratic rainfall event. Environ Microbiol Rep 15:459–483. doi:10.1111/1758-2229.13167.
- Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, McHardy AC, Schulze-Lefert P. 2015. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 17:392–403. doi:10.1016/j.chom.2015.01.011.
- Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. doi:10.1146/annurev-arplant-050312-120106.
- Calabrese S, Cusant L, Sarazin A, Niehl A, Erban A, Brulé D, Recorbet G, Wipd D, Roux C, Kopka J, et al. 2019. Imbalanced regulation of fungal nutrient transports according to phosphate availability in a symbiocosm formed by poplar, sorghum, and Rhizophagus irregularis. Front Plant Sci. 10:1617. doi:10.3389/fpls.2019.01617.
- Casieri L, Ait Lahmidi N, Doidy J, Veneault-Fourrey C, Migeon A, Bonneau L, Courty P-E, Garcia K, Charbonnier M, Delteil A, et al. 2013. Biotrophic transportome in mutualistic plant–fungal interactions. Mycorrhiza 23:597–625. doi:10.1007/s00572-013-0496-9.
- Chagnon P-L, Bradley RL, Maherali H, Klironomos JN. 2013. A trait-based framework to understand life history of mycorrhizal fungi. Trends Plant Sci 18:484–491. doi:10.1016/j.tplants.2013.05.001.
- Chai YN, Schachtman DP. 2022. Root exudates impact plant performance under abiotic stress. Trends Plant Sci 27:80–91. doi:10.1016/j.tplants.2021.08.003.
- Chen M, Arato M, Borghi L, Nouri E, Reinhardt D. 2018. Beneficial services of arbuscular mycorrhizal fungi – from ecology to application. Front Plant Sci 9:1270. doi:10.3389/fpls.2018.01270.
- Cheng S, Zou YN, Kuča K, Hashem A, Abd Allah EF, Wu QS. 2021. Elucidating the mechanisms underlying enhanced drought tolerance in plants mediated by arbuscular mycorrhizal fungi. Front Microbiol 12:809473. doi:10.3389/fmicb.2021.809473.
- Chitarra W, Pagliarani C, Maserti B, Lumini E, Siciliano I, Cascone P, Schubert A, Gambino G, Balestrini R, Guerrieri E. 2016. Insights on the impact of arbuscular mycorrhizal symbiosis on tomato tolerance to water stress. Plant Physiol 171:1009-1023. doi:10.1104/pp.16.00307.
- Choi J, Summers W, Paszkowski U. 2018. Mechanisms underlying establishment of arbuscular mycorrhizal symbioses. Annu Rev Phytopathol 56:135–160. doi:10.1146/annurev-phyto-080516-035521.
- Corona Ramírez A, Symanczik S, Gallusser T, Bodenhausen N. 2023. Quantification of arbuscular mycorrhizal fungi root colonization in wheat, tomato, and leek using absolute qPCR. Mycorrhiza 33:387–397. doi:10.1007/s00572-023-01122-8.
- Defrenne CE, Childs J, Fernandez CW, Taggart M, Nettles WR, Allen MF, Hanson PJ, Iversen CM. 2020. High-resolution minirhizotrons advance our understanding of root-fungal dynamics in an experimentally warmed peatland. Plants people planet. 3:640–652. doi:10.1002/ppp3.10172.
- Deguchi S, Matsuda Y, Takenaka C, Sugiura Y, Ozawa H, Ogata Y. 2017. Proposal of a new estimation method of colonization rate of arbuscular mycorrhizal fungi in the roots of Chengiopanax sciadophylloides. Mycobiology. 45:15–19. doi:10.5941/MYCO.2017.45.1.15
- De La Providencia IE, De Souza FA, Fernández F, Delmas NS, Declerck S. 2004. Arbuscular mycorrhizal fungi reveal distinct patterns of anastomosis formation and hyphal healing mechanisms between different phylogenic groups. New Phytol 165:261–271. doi:10.1111/j.1469-8137.2004.01236.x.
- Dell’Amico J, Torrecillas A, Rodríguez P, Morte A, Sánchez-Blanco MJ. 2002. Responses of tomato plants associated with the arbuscular mycorrhizal fungus Glomus clarum during drought and recovery. J Agric Sci 138:387–393. doi:10.1017/s0021859602002101.
- de Souza RSC, Okura VK, Armanhi JSL, Jorrín B, Lozano N, da Silva MJ, González-Guerrero M, de Araújo LM, Verza NC, Bagheri HC, et al. 2016. Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci Rep 6. doi:10.1038/srep28774.
- de Vries FT, Griffiths RI, Knight CG, Nicolitch O, Williams A. 2020. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science. 368:270–274. doi:10.1126/science.aaz5192.
- de Vries FT, Williams A, Stringer F, Willcocks R, McEwing R, Langridge H, Straathof AL. 2019. Changes in root-exudate-induced respiration reveal a novel mechanism through which drought affects ecosystem carbon cycling. New Phytol 224:132–145. doi:10.1111/nph.16001.
- Djighaly PI, Diagne N, Ngom M, Ngom D, Hocher V, Fall D, Diouf D, Laplaze L, Svistoonoff S, Champion A. 2018. Selection of arbuscular mycorrhizal fungal strains to improve Casuarina equisetifolia L. and Casuarina glauca Sieb. tolerance to salinity. Ann For Sci 75:72. doi:10.1007/s13595-018-0747-1.
- Dohroo A, Sharma DR. 2012. Role of plant growth promoting rhizobacteria, arbuscular mycorrhizal fungi and their helper bacteria on growth parameters and root rot of apple. World J. Food Sci. Technol. 2:35–38.
- Duc NH, Vo AT, Haddidi I, Daood H, Posta K. 2021. Arbuscular mycorrhizal fungi improve tolerance of the medicinal plant Eclipta prostrata (L.) and induce major changes in polyphenol profiles under salt stresses. Front Plant Sci 11. doi:10.3389/fpls.2020.612299.
- Elser JJ, Bracken MES, Cleland EE, Gruner DS, Harpole WS, Hillebrand H, Ngai JT, Seabloom EW, Shurin JB, Smith JE. 2007. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett 10:1135–1142. doi:10.1111/j.1461-0248.2007.01113.x.
- Escudero-Martinez C, Bulgarelli D. 2019. Tracing the evolutionary routes of plant–microbiota interactions. Curr Opin Microbiol 49:34–40. doi:10.1016/j.mib.2019.09.013.
- Essiane-Ondo O, Zerbib J, Gianinazzi S, Wipf D. 2019. Wheat landraces with low mycorrhizing ability at field respond differently to inoculation with artificial or indigenous arbuscular mycorrhizal fungal communities. Symbiosis. 78:229–240. doi:10.1007/s13199-019-00612-8.
- Evangelisti E, Turner C, McDowell A, Shenhav L, Yunusov T, Gavrin A, Servante EK, Quan C, Schornack S. 2021. Deep learning-based quantification of arbuscular mycorrhizal fungi in plant roots. New Phytol 232:2207–2219. doi:10.1111/nph.17697.
- Fan X, Xie H, Huang X, Zhang S, Nie Y, Chen H, Xie X, Tang M. 2023. A module centered on the transcription factor Msn2 from arbuscular mycorrhizal fungus Rhizophagus irregularis regulates drought stress tolerance in the host plant. New Phytol 240:1497–1518. doi:10.1111/nph.19077.
- Fiorilli V, Catoni M, Miozzi L, Novero M, Accotto GP, Lanfranco L. 2009. Global and cell-type gene expression profiles in tomato plants colonized by an arbuscular mycorrhizal fungus. New Phytol 184:975–987. doi:10.1111/j.1469-8137.2009.03031.x.
- Fiorilli V, Maghrebi M, Novero M, Votta C, Mazzarella T, Buffoni B, Astolfi S, Vigani G. 2022. Arbuscular mycorrhizal symbiosis differentially affects the nutritional status of two durum wheat genotypes under drought conditions. Plants. 11:804. doi:10.3390/plants11060804.
- Fitter AH, Graves JD, Watkins NK, Robinson D, Scrimgeour C. 1998. Carbon transfer between plants and its control in networks of arbuscular mycorrhizas. Funct Ecol 12:406–412. doi:10.1046/j.1365-2435.1998.00206.x.
- Fontaine S, Abbadie L, Aubert M, Barot S, Bloor JMG, Derrien D, Duchene O, Gross N, Henneron L, Le Roux X, et al. 2023. Plant–soil synchrony in nutrient cycles: learning from ecosystems to design sustainable agrosystems. Glob Chang Biol 30. doi:10.1111/gcb.17034.
- Francis R, Read DJ. 1984. Direct transfer of carbon between plants connected by vesicular–arbuscular mycorrhizal mycelium. Nature. 307:53–56. doi:10.1038/307053a0.
- Freschet GT, Violle C, Bourget MY, Scherer-Lorenzen M, Fort F. 2018. Allocation, morphology, physiology, architecture: the multiple facets of plant above- and below-ground responses to resource stress. New Phytol 219:1338–1352. doi:10.1111/nph.15225.
- Galindo-Castañeda T, Lynch JP, Six J, Hartmann M. 2022. Improving soil resource uptake by plants through capitalizing on synergies between root architecture and anatomy and root-associated microorganisms. Front Plant Sci 13. doi:10.3389/fpls.2022.827369.
- Gamalero E, Berta G, Massa N, Glick B R, Lingua G. 2008. Synergistic interactions between the ACC deaminase-producing bacterium Pseudomonas putida UW4 and the AM fungus Gigaspora rosea positively affect cucumber plant growth. FEMS Microbiol Ecol. 64:459–467. doi:10.1111/j.1574-6941.2008.00485.x.
- Gamalero E, Trotta A, Massa N, Copetta A, Martinotti MG, Berta G. 2003. Impact of two fluorescent pseudomonads and an arbuscular mycorrhizal fungus on tomato plant growth, root architecture and P acquisition. Mycorrhiza 14:185–192. doi:10.1007/s00572-003-0256-3.
- Garcia K, Doidy J, Zimmermann SD, Wipf D, Courty P-E. 2016. Take a trip through the plant and fungal transportome of mycorrhiza. Trends Plant Sci. 21:937–950. doi:10.1016/j.tplants.2016.07.010.
- Gianinazzi S, Gollotte A, Binet M-N, van Tuinen D, Redecker D, Wipf D. 2010. Agroecology: the key role of arbuscular mycorrhizas in ecosystem services. Mycorrhiza. 20:519–530. doi:10.1007/s00572-010-0333-3.
- Gioia T, Galinski A, Lenz H, Müller C, Lentz J, Heinz K, Briese C, Putz A, Fiorani F, Watt M, et al. 2016. GrowScreen-PaGe, a non-invasive, high-throughput phenotyping system based on germination paper to quantify crop phenotypic diversity and plasticity of root traits under varying nutrient supply. Funct Plant Biol 44:76–93. doi:10.1071/FP16128.
- Giovannetti M, Fortuna P, Citernesi AS, Morini S, Nuti MP. 2001. The occurrence of anastomosis formation and nuclear exchange in intact arbuscular mycorrhizal networks. New Phytol 151:717–724. doi:10.1046/j.0028-646x.2001.00216.x.
- Giovannetti M, Mosse B. 1980. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol 84:489–500. doi:10.1111/j.1469-8137.1980.tb04556.x.
- Goddard M-L, Belval L, Martin IR, Roth L, Laloue H, Deglène-Benbrahim L, Valat L, Bertsch C, Chong J. 2021. Arbuscular mycorrhizal symbiosis triggers major changes in primary metabolism together with modification of defense responses and signaling in both roots and leaves of Vitis vinifera. Front Plant Sci 12. doi:10.3389/fpls.2021.721614.
- Gollotte A, van Tuinen D, Atkinson D. 2003. Diversity of arbuscular mycorrhizal fungi colonising roots of the grass species Agrostis capillaris and Lolium perenne in a field experiment. Mycorrhiza 14:111–117. doi:10.1007/s00572-003-0244-7.
- González Guzmán M, Cellini F, Fotopoulos V, Balestrini R, Arbona V. 2021. New approaches to improve crop tolerance to biotic and abiotic stresses. Physiol Plant 174. doi:10.1111/ppl.13547.
- Grayson M. 2013. Agriculture and drought. Nature. 501:S1. doi:10.1038/501s1a.
- Guether M, Neuhäuser B, Balestrini R, Dynowski M, Ludewig U, Bonfante P. 2009. A mycorrhizal-specific ammonium transporter from Lotus japonicus acquires nitrogen released by arbuscular mycorrhizal fungi. Plant Physiol 150:73–83. doi:10.1104/pp.109.136390.
- Gupta A, Rico-Medina A, Caño-Delgado AI. 2020. The physiology of plant responses to drought. Science. 368:266–269. doi:10.1126/science.aaz7614.
- Hart MM, Reader RJ. 2002. Taxonomic basis for variation in the colonization strategy of arbuscular mycorrhizal fungi. New Phytol 153:335–344. doi:10.1046/j.0028-646x.2001.00312.x.
- Helber N, Wippel K, Sauer N, Schaarschmidt S, Hause B, Requena N. 2011. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell. 23:3812–3823. doi:10.1105/tpc.111.089813.
- Hewins CR, Carrino-Kyker SR, Burke DJ. 2015. Seasonal variation in mycorrhizal fungi colonizing roots of Allium tricoccum (wild leek) in a mature mixed hardwood forest. Mycorrhiza 25:469–483. doi:10.1007/s00572-015-0628-5.
- Hou S, Wolinska KW, Hacquard S. 2021. Microbiota-root-shoot-environment axis and stress tolerance in plants. Curr Opin Plant Biol 62:102028. doi:10.1016/j.pbi.2021.102028.
- Huang X-F, Chaparro JM, Reardon KF, Zhang R, Shen Q, Vivanco JM. 2014. Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany. 92:267–275. doi:10.1139/cjb-2013-0225.
- Iannucci A, Fragasso M, Beleggia R, Nigro F, Papa R. 2017. Evolution of the crop rhizosphere: impact of domestication on root exudates in tetraploid wheat (Triticum turgidum L.). Front Plant Sci 8. doi:10.3389/fpls.2017.02124.
- Jeudy C, Adrian M, Baussard C, Bernard C, Bernaud E, Bourion V, Busset H, Cabrera-Bosquet L, Cointault F, Han S, et al. 2016. Rhizotubes as a new tool for high throughput imaging of plant root development and architecture: test, comparison with pot grown plants and validation. Plant Methods. 12. doi:10.1186/s13007-016-0131-9.
- Johnson MG, Meyer PF. 1998. Mechanical advancing handle that simplifies minirhizotron camera registration and image collection. J Environ Qual 27:710–714. doi:10.2134/jeq1998.00472425002700030031x.
- Jubery TZ, Carley CN, Singh A, Sarkar S, Ganapathysubramanian B, Singh AK. 2021. Using machine learning to develop a fully automated soybean nodule acquisition pipeline (snap). Plant Phenomics. 2021:9834746. doi:10.34133/2021/9834746.
- Keller-Pearson M, Bortolazzo A, Willems L, Smith B, Peterson A, Ané J-M, Silva E. 2023. A dual transcriptomic approach reveals contrasting patterns of differential gene expression during drought in arbuscular mycorrhizal fungus and carrot. MPMI. 36:821–832. doi:10.1094/mpmi-04-23-0038-r.
- Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, Fellbaum CR, Kowalchuk GA, Hart MM, Bago A, et al. 2011. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science. 333:880–882. doi:10.1126/science.1208473.
- Kinnunen-Grubb M, Sapkota R, Vignola M, Nunes IM, Nicolaisen M. 2020. Breeding selection imposed a differential selective pressure on the wheat root-associated microbiome. FEMS Microbiol. Ecol. 96:fiaa196. doi:10.1093/femsec/fiaa196.
- Koch AM, Antunes PM, Maherali H, Hart MM, Klironomos JN. 2017. Evolutionary asymmetry in the arbuscular mycorrhizal symbiosis: conservatism in fungal morphology does not predict host plant growth. New Phytol 214:1330–1337. doi:10.1111/nph.14465.
- Kumar A, Lin H, Li Q, Ruan Y, Cousins D, Li F, Gao S, Jackson K, Wen J, Murray JD, Xu P. 2022. Anthocyanin pigmentation as a quantitative visual marker for arbuscular mycorrhizal fungal colonization of Medicago truncatula roots. New Phytol 236:988–1998. doi:10.1111/nph.18504.
- Lareen A, Burton F, Schäfer P. 2016. Plant root-microbe communication in shaping root microbiomes. Plant Mol Biol 90:575–587. doi:10.1007/s11103-015-0417-8.
- Lesk C, Rowhani P, Ramankutty N. 2016. Influence of extreme weather disasters on global crop production. Nature. 529:84–87. doi:10.1038/nature16467.
- Ling F, Su Q, Jiang H, Cui J, He X, Wu Z, Zhang Z, Liu J, Zhao Y. 2020. Effects of strigolactone on photosynthetic and physiological characteristics in salt-stressed rice seedlings. Sci Rep 10:6183. doi:10.1038/s41598-020-63352-6.
- Lu T, Ke M, Lavoie M, Jin Y, Fan X, Zhang Z, Fu Z, Sun L, Gillings M, Peñuelas J, et al. 2018. Rhizosphere microorganisms can influence the timing of plant flowering. Microbiome. 6:231. doi:10.1186/s40168-018-0615-0.
- Lynch JP. 2013. Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann Bot 112:347–357. doi:10.1093/aob/mcs293.
- Lynch JP, Strock CF, Schneider HM, Sidhu JS, Ajmera I, Galindo-Castañeda T, Klein SP, Hanlon MT. 2021. Root anatomy and soil resource capture. Plant Soil. 466:21–63. doi:10.1007/s11104-021-05010-y.
- Ma S, Zhu L, Wang J, Liu X, Jia Z, Li C, Liu J, Zeng J, Zhang J. 2022. Arbuscular mycorrhizal fungi promote Gleditsia sinensis Lam. root growth under salt stress by regulating nutrient uptake and physiology. Forests. 13:688. doi:10.3390/f13050688.
- Mahoney AK, Yin C, Hulbert SH. 2017. Community structure, species variation, and potential functions of rhizosphere-associated bacteria of different winter wheat (Triticum aestivum) cultivars. Front Plant Sci 8:132. doi:10.3389/fpls.2017.00132.
- Marro N, Grilli G, Soteras F, Caccia M, Longo S, Cofré N, Borda V, Burni M, Janoušková M, Urcelay C. 2022. The effects of arbuscular mycorrhizal fungal species and taxonomic groups on stressed and unstressed plants: a global meta-analysis. New Phytol 235:320–332. doi:10.1111/nph.18102.
- Mesny F, Hacquard S, Thomma BP. 2023. Co-evolution within the plant holobiont drives host performance. EMBO Rep 24. doi:10.15252/embr.202357455.
- Mikkelsen BL, Rosendahl S, Jakobsen I. 2008. Underground resource allocation between individual networks of mycorrhizal fungi. New Phytol 180:890–898. doi:10.1111/j.1469-8137.2008.02623.x.
- Montero H, Choi J, Paszkowski U. 2018. Arbuscular mycorrhizal phenotyping: the dos and don’ts. New Phytol 221:1182–1186. doi:10.1111/nph.15489.
- Moriondo M, Giannakopoulos C, Bindi M. 2010. Climate change impact assessment: the role of climate extremes in crop yield simulation. Clim Change 104:679–701. doi:10.1007/s10584-010-9871-0.
- Muta K, Takata S, Utsumi Y, Matsumura A, Iwamura M, Kise K. 2022. TAIM: tool for analyzing root images to calculate the infection rate of arbuscular mycorrhizal fungi. Front Plant Sci 13:881382. doi:10.3389/fpls.2022.881382.
- Nerva L, Sandrini M, Moffa L, Velasco R, Balestrini R, Chitarra W. 2022. Breeding toward improved ecological plant–microbiome interactions. Trends Plant Sci 27:1134–1143. doi:10.1016/j.tplants.2022.06.004.
- Newman EI, Devoy CLN, Easen NJ, Fowles KJ. 1994. Plant species that can be linked by VA mycorrhizal fungi. New Phytol 126:691–693. doi:10.1111/j.1469-8137.1994.tb02963.x.
- Noceto P-A, Bettenfeld P, Boussageon R, Hériché M, Sportes A, van Tuinen D, Courty P-E, Wipf D. 2021. Arbuscular mycorrhizal fungi, a key symbiosis in the development of quality traits in crop production, alone or combined with plant growth-promoting bacteria. Mycorrhiza. 31:655–669. doi:10.1007/s00572-021-01054-1.
- Noceto P-A, Durney C, van Tuinen D, de Sousa J, Wipf D, Courty P-E. 2023. Arbuscular mycorrhizal fungal communities differ in neighboring vineyards of different ages. Mycorrhiza. 33:241–248. doi:10.1007/s00572-023-01117-5.
- Ordoñez YM, Fernandez BR, Lara LS, Rodriguez A, Uribe-Vélez D, Sanders IR. 2016. Bacteria with phosphate solubilizing capacity alter mycorrhizal fungal growth both inside and outside the root and in the presence of native microbial communities. PLoS One. 11:e0154438. doi:10.1371/journal.pone.0154438.
- Ordookhani K, Zare M. 2011. Effect of Pseudomonas, Azotobacter and arbuscular mycorrhiza fungi on lycopene, antioxidant activity and total soluble solid in tomato (Lycopersicon esculentum F1 Hybrid, Delba). Adv. Environ. Biol. 5:1290–1294.
- Pandey A, Sharma M, Pandey GK. 2016. Emerging roles of strigolactones in plant responses to stress and development. Front Plant Sci 7:434. doi:10.3389/fpls.2016.00434.
- Pedersen O, Perata P, Voesenek LACJ. 2017. Flooding and low oxygen responses in plants. Funct Plant Biol 44:iii. doi:10.1071/fpv44n9_fo.
- Pérez-Jaramillo JE, Carrión VJ, Bosse M, Ferrão LFV, de Hollander M, Garcia AAF, Ramírez CA, Mendes R, Raaijmakers JM. 2017. Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J 11:2244–2257. doi:10.1038/ismej.2017.85.
- Pérez-Jaramillo JE, de Hollander M, Ramírez CA, Mendes R, Raaijmakers JM, Carrión VJ. 2019. Deciphering rhizosphere microbiome assembly of wild and modern common bean (Phaseolus vulgaris) in native and agricultural soils from Colombia. Microbiome. 7(1). doi:10.1186/s40168-019-0727-1.
- Pfeifer J, Kirchgessner N, Colombi T, Walter A. 2015. Rapid phenotyping of crop root systems in undisturbed field soils using X-ray computed tomography. Plant Methods. 11:41. doi:10.1186/s13007-015-0084-4.
- Pflugfelder D, Metzner R, van Dusschoten D, Reichel R, Jahnke S, Koller R. 2017. Non-invasive imaging of plant roots in different soils using magnetic resonance imaging (MRI). Plant Methods. 13:102. doi:10.1186/s13007-017-0252-9.
- Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. 2013. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789–799. doi:10.1038/nrmicro3109.
- Pimprikar P, Carbonnel S, Paries M, Katzer K, Klingl V, Bohmer MJ, Karl L, Floss DS, Harrison MJ, Parniske M, Gutjahr C. 2016. A CCAMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr Biol 26:1126. doi:10.1016/j.cub.2016.04.021.
- Pimprikar P, Gutjahr C. 2018. Transcriptional regulation of arbuscular mycorrhiza development. Plant Cell Physiol 59:678–695. doi:10.1093/pcp/pcy024.
- Pivato B, Offre P, Marchelli S, Barbonaglia B, Mougel C, Lemanceau P, Berta G. 2008. Bacterial effects on arbuscular mycorrhizal fungi and mycorrhiza development as influenced by the bacteria, fungi, and host plant. Mycorrhiza. 19:81–90. doi:10.1007/s00572-008-0205-2.
- Pollastri S, Savvides A, Pesando M, Lumini E, Volpe MG, Ozudogru EA, Faccio A, De Cunzo F, Michelozzi M, Lambardi M, et al. 2017. Impact of two arbuscular mycorrhizal fungi on Arundo donax L. response to salt stress. Planta. 247:573–585. doi:10.1007/s00425-017-2808-3.
- Powell JR, Parrent JL, Hart MM, Klironomos JN, Rillig MC, Maherali H. 2009. Phylogenetic trait conservatism and the evolution of functional trade-offs in arbuscular mycorrhizal fungi. Proc. Royal Soc. B. 276:4237–4245. doi:10.1098/rspb.2009.1015.
- Powell JR, Rillig MC. 2018. Biodiversity of arbuscular mycorrhizal fungi and ecosystem function. New Phytol. 220:1059–1075. doi:10.1111/nph.15119.
- Quiroga G, Erice G, Aroca R, Zamarreño ÁM, García-Mina JM, Ruiz-Lozano JM. 2020. Radial water transport in arbuscular mycorrhizal maize plants under drought stress conditions is affected by indole-acetic acid (IAA) application. J Plant Physiol 246–247:153115. doi:10.1016/j.jplph.2020.153115.
- Recchia GH, Konzen ER, Cassieri F, Caldas DGG, Tsai SM. 2018. Arbuscular mycorrhizal symbiosis leads to differential regulation of drought-responsive genes in tissue-specific root cells of common bean. Front Microbiol 9:1339. doi:10.3389/fmicb.2018.01339.
- Reid CPP, Woods FW. 1969. Translocation of C(14)-labeled compounds in mycorrhizae and it implications in interplant nutrient cycling. Ecology. 50:179–187. doi:10.2307/1934844.
- Remmler L, Clairmont L, Rolland-Lagan A, Guinel FC. 2014. Standardized mapping of nodulation patterns in legume roots. New Phytol. 202:1083–1094. doi:10.1111/nph.12712.
- Rich MK, Nouri E, Courty P-E, Reinhardt D. 2017. Diet of arbuscular mycorrhizal fungi: bread and butter? Trends Plant Sci 22:652–660. doi:10.1016/j.tplants.2017.05.008.
- Rivero J, Álvarez D, Flors V, Azcón-Aguilar C, Pozo MJ. 2018. Root metabolic plasticity underlies functional diversity in mycorrhiza-enhanced stress tolerance in tomato. New Phytol 220:1322–1336. doi:10.1111/nph.15295.
- Roesti D, Ineichen K, Braissant O, Redecker D, Wiemken A, Aragno M. 2005. Bacteria associated with spores of the arbuscular mycorrhizal fungi Glomus geosporum and Glomus constrictum. Applied Env. Microbiol. 71:6673–6679. doi:10.1128/aem.71.11.6673-6679.2005.
- Rolfe SA, Griffiths J, Ton J. 2019. Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr Opin Microbiol 49:73–82. doi:10.1016/j.mib.2019.10.003.
- Rowland DL, Smith C, Cook A, Mason A, Schreffler A, Bennett J. 2015. Visualization of peanut nodules and seasonal nodulation pattern in different tillage systems using a Minirhizotron system. Peanut Science. 42:1–10. doi:10.3146/0095-3679-42.1.1.
- Rúa MA, Antoninka A, Antunes PM, Chaudhary VB, Gehring C, Lamit LJ, Piculell BJ, Bever JD, Zabinski C, Meadow JF, et al. 2016. Home-field advantage? Evidence of local adaptation among plants, soil, and arbuscular mycorrhizal fungi through meta-analysis. BMC Evol Biol 16:122. doi:10.1186/s12862-016-0698-9.
- Ruiz-Lozano JM, Aroca R, Zamarreño ÁM, Molina S, Andreo-Jiménez B, Porcel R, García-Mina JM, Ruyter-Spira C, López-Ráez JA. 2015. Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Environ 39:441–452. doi:10.1111/pce.12631.
- Ruiz-Lozano JM, Azcón R. 1995. Hyphal contribution to water uptake in mycorrhizal plants as affected by the fungal species and water status. Physiol. Plantarum. 95:472–478. doi:10.1111/j.1399-3054.1995.tb00865.x.
- Rush TA, Puech-Pagès V, Bascaules A, Jargeat P, Maillet F, Haouy A, Maës AQ, Carriel CC, Khokhani D, Keller-Pearson M, Tannous J. 2020. Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat Commun 11:3897. doi:10.1038/s41467-020-17615-5.
- Schillaci M, Arsova B, Walker R, Smith PMC, Nagel KA, Roessner U, Watt M. 2020. Time-resolution of the shoot and root growth of the model cereal Brachypodium in response to inoculation with Azospirillum bacteria at low phosphorus and temperature. Plant Growth Regul 93:149–162. doi:10.1007/s10725-020-00675-4.
- Schloter M, Matyssek R. 2009. Tuning growth versus defence–belowground interactions and plant resource allocation. Plant & Soil. 323:1–5. doi:10.1007/s11104-009-0070-6.
- Schmidt JE, Bowles TM, Gaudin ACM. 2016. Using ancient traits to convert soil health into crop yield: impact of selection on maize root and rhizosphere function. Front Plant Sci 7. doi:10.3389/fpls.2016.00373.
- Schneider HM, Lynch JP. 2020. Should root plasticity be a crop breeding target? Front Plant Sci 11:546. doi:10.3389/fpls.2020.00546.
- Sciascia I, Crosino A, Genre A. 2023. Quantifying root colonization by a symbiotic fungus using automated image segmentation and machine learning approaches. Sci Rep. 13:14830. doi:10.1038/s41598-023-39217-z.
- Seethepalli A, Dhakal K, Griffiths M, Guo H, Freschet GT, York LM. 2021. Rhizovision explorer: open-source software for root image analysis and measurement standardization. AoB PLANTS. 13:plab056. doi:10.1093/aobpla/plab056.
- Shi J, Wang X, Wang E. 2023. Mycorrhizal symbiosis in plant growth and stress adaptation: from genes to ecosystems. Annu Rev Plant Biol 74:569–607. doi:10.1146/annurev-arplant-061722-090342.
- Sillo F, Marino G, Franchi E, Haworth M, Zampieri E, Pietrini I, Fusini D, Mennone C, Centritto M, Balestrini R. 2022. Impact of irrigation water deficit on two tomato genotypes grown under open field conditions: from the root-associated microbiota to the stress responses. Ital J Agron 17(3). doi:10.4081/ija.2022.2130.
- Simard SW, Perry DA, Jones MD, Myrold DD, Durall DM, Molina R. 1997. Net transfer of carbon between ectomycorrhizal tree species in the field. Nature. 388:579–582. doi:10.1038/41557.
- Smith SE, Read DJ. 2008. Mycorrhizal symbiosis, 3rd ed. London, UK: Academic Press.
- Sportes A, Hériché M, Boussageon R, Noceto PA, van Tuinen D, Wipf D, Courty PE. 2022. Correction to: a historical perspective on mycorrhizal mutualism emphasizing arbuscular mycorrhizas and their emerging challenges. Mycorrhiza. 32:497–497. doi:10.1007/s00572-022-01094-1.
- Stearns JC, Woody OZ, McConkey BJ, Glick BR. 2012. Effects of bacterial ACC deaminase on Brassica napus gene expression. MPMI. 25:668–676. doi:10.1094/mpmi-08-11-0213.
- Subramanian KS, Santhanakrishnan P, Balasubramanian P. 2006. Responses of field grown tomato plants to arbuscular mycorrhizal fungal colonization under varying intensities of drought stress. Sci Hortic 107:245–253. doi:10.1016/j.scienta.2005.07.006.
- Sultan SE. 2000. Phenotypic plasticity for plant development, function and life history. Trends Plant Sci 5:537–542. doi:10.1016/s1360-1385(00)01797-0.
- Sun L, Ataka M, Han M, Han Y, Gan D, Xu T, Guo Y, Zhu B. 2020. Root exudation as a major competitive fine-root functional trait of 18 coexisting species in a subtropical forest. New Phytol 229:259–271. doi:10.1111/nph.16865.
- Sun Z, Song J, Xin X, Xie X, Zhao B. 2018. Arbuscular mycorrhizal fungal 14-3-3 proteins are involved in arbuscule formation and responses to abiotic stresses during AM symbiosis. Front Microbiol 9:91. doi:10.3389/fmicb.2018.00091.
- Svenningsen NB, Watts-Williams SJ, Joner EJ, Battini F, Efthymiou A, Cruz-Paredes C, Nybroe O, Jakobsen I. 2018. Suppression of the activity of arbuscular mycorrhizal fungi by the soil microbiota. ISME J 12:1296–1307. doi:10.1038/s41396-018-0059-3.
- Symanczik S, Courty PE, Boller T, Wiemken A, Al-Yahya’ei MN. 2015. Impact of water regimes on an experimental community of four desert arbuscular mycorrhizal fungal (AMF) species, as affected by the introduction of a non-native AMF species. Mycorrhiza. 25:639–647. doi:10.1007/s00572-015-0638-3.
- Symanczik S, Kruetzmann J, Nehls U, Boller T, Courty PE. 2020. Expression of major intrinsic protein genes in Sorghum bicolor roots under water deficit depends on arbuscular mycorrhizal fungal species. Soil Biol Biochem 140:107643. doi:10.1016/j.soilbio.2019.107643.
- Symanczik S, Lehmann MF, Wiemken A, Boller T, Courty P-E. 2018. Effects of two contrasted arbuscular mycorrhizal fungal isolates on nutrient uptake by Sorghum bicolor under drought. Mycorrhiza. 28:779–785. doi:10.1007/s00572-018-0853-9.
- Tavasolee A, Aliasgharzad N, SalehiJouzani G, Mardi M, Asgharzadeh A. 2013. Interactive effects of arbuscular mycorrhizal fungi and rhizobial strains on chickpea growth and nutrient content in plant. African J. Biotech. 10:7585–7591.
- Taylor HM. 1987. Minirhizotron observation tubes: Methods and applications for measuring rhizosphere dynamics. American Society of Agronomy, Madison Wisconsin. ASA Special Publication Number 50.
- Thoms D, Liang Y, Haney CH. 2021. Maintaining symbiotic homeostasis: how do plants engage with beneficial microorganisms while at the same time restricting pathogens? MPMI. 34:462–469. doi:10.1094/mpmi-11-20-0318-fi.
- Thonar C, Erb A, Jansa J. 2011. Real-time PCR to quantify composition of arbuscular mycorrhizal fungal communities—marker design, verification, calibration and field validation. Mol. Ecol. Res. 12:219–232. doi:10.1111/j.1755-0998.2011.03086.x.
- Timoneda A, Yunusov T, Quan C, Gavrin A, Brockington SF, Schornack S. 2021. Mycored: betalain pigments enable in vivo real-time visualisation of arbuscular mycorrhizal colonisation. PLoS Biol 19:e3001326. doi:10.1371/journal.pbio.3001326.
- Todeschini V, AitLahmidi N, Mazzucco E, Marsano F, Gosetti F, Robotti E, Bona E, Massa N, Bonneau L, Marengo E, et al. 2018. Impact of beneficial microorganisms on strawberry growth, fruit production, nutritional quality, and volatilome. Front Plant Sci 9:1611. doi:10.3389/fpls.2018.01611.
- Trouvelot A, Kough JL, Gianinazzi-Pearson V. 1986. Mesure du taux de mycorhization VA dun système radiculaire: recherche de méthodes d’estimation ayant une signification fonctionnelle. In: Gianinazzi-Pearson V., Gianinazzi S., editor. Physiological and Genetical Aspects of Mycorrhizae. INRA Press; p. 217–221.
- Tsuzuki S, Handa Y, Takeda N, Kawaguchi M. 2016. Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. MPMI. 29:277–286. doi:10.1094/mpmi-10-15-0234-r.
- Valente J, Gerin F, Le Gouis J, Moënne-Loccoz Y, Prigent–Combaret C. 2019. Ancient wheat varieties have a higher ability to interact with plant growth-promoting rhizobacteria. Plant Cell Envir. 43:246–260. doi:10.1111/pce.13652.
- van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, Wiemken A, Sanders IR. 1998. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature. 396:69–72. doi:10.1038/23932.
- Vangelisti A, Natali L, Bernardi R, Sbrana C, Turrini A, Hassani-Pak K, Hughes D, Cavallini A, Giovannetti M, Giordani T. 2018. Transcriptome changes induced by arbuscular mycorrhizal fungi in sunflower (Helianthus annuus L.) roots. Sci Rep 8:4. doi:10.1038/s41598-017-18445-0.
- Vierheilig H, Schweiger P, Brundrett M. 2005. An overview of methods for the detection and observation of arbuscular mycorrhizal fungi in roots. Physiol. Plantarum. 125:393–404. doi:10.1111/j.1399-3054.2005.00564.x.
- Vivas A, Barea JM, Azcón R. 2005. Interactive effect of Brevibacillus brevis and Glomus mosseae, both isolated from Cd contaminated soil, on plant growth, physiological mycorrhizal fungal characteristics and soil enzymatic activities in Cd polluted soil. Environ Pollut 134:257–266. doi:10.1016/j.envpol.2004.07.029.
- Volpe V, Chialva M, Mazzarella T, Crosino A, Capitanio S, Costamagna L, Kohlen W, Genre A. 2023. Long-lasting impact of chitooligosaccharide application on strigolactone biosynthesis and fungal accommodation promotes arbuscular mycorrhiza in Medicago truncatula. New Phytol 237:2316–2331. doi:10.1111/nph.18697.
- Volpe V, Chitarra W, Cascone P, Volpe MG, Bartolini P, Moneti G, Pieraccini G, Di Serio C, Maserti B, Guerrieri E, Balestrini R. 2018. The association with two different arbuscular mycorrhizal fungi differently affects water stress tolerance in tomato. Front Plant Sci. 9:1480. doi:10.3389/fpls.2018.01480.
- Voříšková A, Jansa J, Püschel D, Krüger M, Cajthaml T, Vosátka M, Janoušková M. 2017. Real-time PCR quantification of arbuscular mycorrhizal fungi: does the use of nuclear or mitochondrial markers make a difference? Mycorrhiza. 27:577–585. doi:10.1007/s00572-017-0777-9.
- Vosatka M, Gryndler M, Prikryl Z. 1992. Effect of the rhizosphere bacterium Pseudomonas putida, arbuscular mycorrhizal fungi and substrate composition on the growth of strawberry. Agronomie. 12:859–863. doi:10.1051/agro:19921021.
- Walder F, Brulé D, Koegel S, Wiemken A, Boller T, Courty PE. 2015. Plant phosphorus acquisition in a common mycorrhizal network: regulation of phosphate transporter genes of the Pht1 family in sorghum and flax. New Phytol 205:1632–1645. doi:10.1111/nph.13292.
- Wang F, Zhang L, Zhou J, Rengel Z, George TS, Feng G. 2022. Exploring the secrets of hyphosphere of arbuscular mycorrhizal fungi: processes and ecological functions. Plant & Soil. 481:1–22. doi:10.1007/s11104-022-05621-z.
- Wang Y, Li T, Li Y, Björn L O, Rosendahl S, Olsson P A, Li S, Fu X. 2015. Community dynamics of arbuscular mycorrhizal fungi in high-input and intensively irrigated rice cultivation systems. Appl. Environ. Microbiol. 81:2958–2965. doi:10.1128/AEM.03769-14.
- Waters MT, Gutjahr C, Bennett T, Nelson DC. 2017. Strigolactone signaling and evolution. Ann. Rev. Plant Biol. 68:291–322. doi:10.1146/annurev-arplant-042916-040925.
- Wen T, Yu G-H, Hong W-D, Yuan J, Niu G-Q, Xie P-H, Sun F-S, Guo L-D, Kuzyakov Y, Shen Q-R. 2022. Root exudate chemistry affects soil carbon mobilization via microbial community reassembly. Fundam. Res. 2:697–707. doi:10.1016/j.fmre.2021.12.016.
- Wipf D, Krajinski F, van Tuinen D, Recorbet G, Courty P. 2019. Trading on the arbuscular mycorrhiza market: from arbuscules to common mycorrhizal networks. New Phytol 223:1127–1142. doi:10.1111/nph.15775.
- Xie ZP, Staehelin C, Vierheilig H, Wiemken A, Jabbouri S, Broughton WJ, Vogeli-Lange R, Boller T. 1995. Rhizobial nodulation factors stimulate mycorrhizal colonization of nodulating and nonnodulating soybeans. Plant Physiol 108:1519–1525. doi:10.1104/pp.108.4.1519.
- Xiong J, Lu J, Li X, Qiu Q, Chen J, Yan C. 2021. Effect of rice (Oryza sativa L.) genotype on yield: evidence from recruiting spatially consistent rhizosphere microbiome. Soil Biol Biochem 161:108395. doi:10.1016/j.soilbio.2021.108395.
- Yue H, Yue W, Jiao S, Kim H, Lee Y-H, Wei G, Song W, Shu D. 2023. Plant domestication shapes rhizosphere microbiome assembly and metabolic functions. Microbiome. 11:70. doi:10.1186/s40168-023-01513-1.
- Zappala S, Helliwell JR, Tracy SR, Mairhofer S, Sturrock CJ, Pridmore T, Bennett M, Mooney SJ. 2013. Effects of X-ray dose on rhizosphere studies using X-ray computed tomography. PLoS One. 8:e67250. doi:10.1371/journal.pone.0067250.
- Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi S, Cho H, Karaoz U, Loqué D, Bowen BP, et al. 2018. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3:470–480. doi:10.1038/s41564-018-0129-3.
- Zhang Y, Gao W, Luan H, Tang J, Li R, Li M, Zhang H, Huang S. 2022. Effects of a decade of organic fertilizer substitution on vegetable yield and soil phosphorus pools, phosphatase activities, and the microbial community in a greenhouse vegetable production system. J. Integr. Agric. 21:2119–2133. doi:10.1016/s2095-3119(21)63715-2.
- Zhang Y, Yao Q, Li J, Wang Y, Liu X, Hu Y, Chen J. 2015. Contributions of an arbuscular mycorrhizal fungus to growth and physiology of loquat (Eriobotrya japonica) plants subjected to drought stress. Mycol Prog 14:84. doi:10.1007/s11557-015-1108-1.
- Zheng Y, Xu Z, Liu H, Liu Y, Zhou Y, Meng C, Ma S, Xie Z, Li Y, Zhang CS. 2021. Patterns in the microbial community of salt-tolerant plants and the functional genes associated with salt stress alleviation. Microbiol Spectr 9:e0076721. doi:10.1128/Spectrum.00767-21.

