Abstract
The effects of long-term chronic exposure of human lung cells to multi-walled carbon nanotubes (MWCNT) and their impact upon cellular proteins and lipids were investigated. Since the lung is the major target organ, an in vitro normal bronchial epithelial cell line model was used. Additionally, to better mimic exposure to manufactured nanomaterials at occupational settings, cells were continuously exposed to two non-toxic and low doses of a MWCNT for 13-weeks. MWCNT-treatment increased ROS levels in cells without increasing oxidative DNA damage and resulted in differential expression of multiple anti- and pro-apoptotic proteins. The proteomic analysis of the MWCNT-exposed cells showed that among more than 5000 identified proteins; more than 200 were differentially expressed in the treated cells. Functional analyses revealed association of these differentially regulated proteins to cellular processes such as cell death and survival, cellular assembly, and organization. Similarly, shotgun lipidomic profiling revealed accumulation of multiple lipid classes. Our results indicate that long-term MWCNT-exposure of human normal lung cells at occupationally relevant low-doses may alter both the proteome and the lipidome profiles of the target epithelial cells in the lung.
Introduction
Carbon nanotubes (CNTs) are made of one (single-walled, SWCNT) or multiple (multi-walled, MWCNT) layers of graphene (Iijima Citation1991). CNTs possess unique material properties and are attractive for multiple commercial applications such as electronics, photonics, textiles, renewable energy, construction materials, and in the biomedical and pharmaceutical sectors (De Volder et al. Citation2013). Due to their diverse commercial applications, it is, therefore, estimated that the global market value of CNTs will reach several hundred million USD by 2020 (Jia and Wei Citation2017). This also implies that an increasing number of workers will be exposed to these engineered nanomaterials at various stages of design, production, use, and waste disposal.
The high aspect ratio, fiber-like structure, and observations that exposure to CNTs could result in unintended asbestos-like effects have raised human health concerns (Poland et al. Citation2008; Donaldson et al. Citation2013). Noteworthy, animals exposed to MWCNT suffered from pulmonary inflammation, fibrosis, mesothelioma, and lung adenocarcinoma (Muller et al. Citation2005; Xu et al. Citation2012; Rittinghausen et al. Citation2014; Sargent et al. Citation2014; Kasai et al. Citation2015; Polimeni et al. Citation2015). In some in vitro studies, MWCNT exposure increased reactive oxygen species (ROS), triggered DNA damage, altered cell cycle, and induced apoptosis (Ravichandran et al. Citation2010; Srivastava et al. Citation2011; Wang et al. Citation2012; Cao et al. Citation2014a; Wang et al. Citation2014; Cao et al. Citation2016).
Molecular mechanisms underlying the CNT-induced toxicological endpoints are poorly understood and may include transcriptomic and epigenetic changes. MWCNT exposure may also affect the proteomic profile of the cells. For instance, human epidermal keratinocytes displayed differential expression of more than 30 and 100 proteins after 24 and 48 h of MWCNT exposure, respectively (Witzmann and Monteiro-Riviere Citation2006). Similarly, other proteomic analyses have identified alterations in the expression of several proteins upon MWCNT exposure (Haniu et al. Citation2010; Ju et al. Citation2014; Hilton et al. Citation2015, Citation2017). These findings indicate that toxicoproteomic analyses may provide valuable information on the mechanisms underlying toxicity of MWCNTs. Proteomic studies are scarce within the field of nanomaterials, especially studies addressing the effects of long-term and chronic exposures. This is a potential drawback because harmful effects from MWCNT exposure occur most likely from long-term and repeated exposures at work or during use. There is an increasing need for chronic exposure studies to correlate to such situations. Studies investigating the lipid profiles of cells following MWCNT exposure are also lacking. Lipids are important mediators of physiological responses and are involved in various biological processes and may modify the adverse effects of MWCNT (Atilla-Gokcumen et al. Citation2014; Muro et al. Citation2014).
The aim of this study was to investigate molecular mechanisms associated with long-term but low-dose exposure to a well characterized MWCNT. Toxicoproteomic analysis of pulmonary effects of MWCNT has shown changes in proteins in bronchoalveolar lavage fluid of the exposed mice (Hilton et al. Citation2015). We thus hypothesized that monitoring proteomic changes associated with chronic exposure of the human lung cells might reveal mechanisms contributing to the adverse effects of MWCNT. The lipid profiles of the cells were also studied to investigate involvement of lipids in the adverse effects of CNTs.
Materials and methods
Full description of materials and methods is given in the online Supplementary file.
The MWCNT NM400 characterization
The CNT used in this study is a MWCNT (designated as NM400) obtained from the repository of nanomaterials of European Union’s Joint Research Centre (JRC) and has a mean length of 846 ± 446 nm; a thickness of 11 ± 3 nm; a surface area of 254 g/m3; with a dustiness <420 mg/kg. The detailed physical and chemical properties of NM400 CNT are found at http://publications.jrc.ec.europa.eu/repository/bitstr-eam/JRC91205/mwcnt-online.pdf. We have furthermore measured the diameter of the particles in solution using Dynamic Light Scattering (DLS) which are shown in the Supplementary Figure S3 (online Supplementary Materials and methods section). Zeta potential (surface charge) is believed not to be highly relevant for MWCNTs and therefore was not assessed. It should be noted that size of particles measured with DLS in dispersion and cell culture media determined at 0 h and 72 h are not comparable with Transmission Electron Microscopy (TEM) measurements (Figure S3).
For dispersion and exposure, the NANOGENOTOX dispersion protocol was applied as described previously (Jensen et al. Citation2011). A 2.56 mg/mL stock was prepared in the dispersion medium (DM). The hydrodynamic diameter of the nanoparticles was measured by DLS and further characterization was performed by TEM in the cell culture medium.
Long-term chronic exposure of cells to NM400
Human bronchial epithelial 3-KT (HBEC-3KT) cells used for the long-term chronic exposure experiments were exposed to two concentrations of the NM400, 1.92 µg/cm2 (designated as high dose), and 0.96 µg/cm2 (designed as low dose), with biological duplicates for each exposure. There are no harmonized occupational exposure limits (OEL), and doses in this study are comparable with recommended exposure limit (REL) of 1 µg/m3 proposed by NIOSH, USA (https://www.cdc.gov/niosh/docs/2013-145/pdfs/2013-145.pdf). Control cells were exposed only to DM. Briefly, 2.5 × 105 cells/15-cm dish were plated in duplicates on day 1 and exposed on days 3 and 5 to freshly prepared NM400 CNT and incubated for another 72 h. After this, the cell viability was assessed by trypan blue assay and cells were subcultured for new rounds of exposures. This procedure was repeated for 13 weeks and then samples were prepared for proteomics and lipidomics analyses. Cellular uptake of MWCNTs and their presence in cytoplasm and phagosomes of human lung epithelial cells has been documented in previous studies using TEM (Snyder et al. Citation2014; Ursini et al. Citation2014). Based on these studies, we chose not to focus on cellular uptake of MWCNT by TEM. However, laser scanning confocal microscopy (LSM) showed presence of CNTs in the cells (Supplementary Figure S2).
Analysis of ROS production using 2′,7′-dichlorofluorescin diacetate (DCF-DA) assay
ROS was measured using DCF-DA assay. Cells were seeded on 6-well plates (3 × 104 cells/well) and incubated overnight, and then exposed to two concentrations (High dose; 1.92 µg/cm2 and low dose; 0.96 µg/cm2) of NM400 for 72 h. After end exposure, 100 µM of DCFDA solution in serum-free medium was added to cells and incubated at 37 °C for 1 h. The cells were then scraped, sonicated for 2 × 10 s in microtubes, centrifuged and supernatants transferred to a 96-well plate in triplicates. Fluorescence was measured at excitation 488 nm and emission 535 nm. Protein concentrations were measured using the Bradford assay and ROS levels were related to total protein. Each experiment was repeated twice using three replicates. Statistical analysis was performed using a paired t-test and p < 0.05 was considered statistically significant. Additionally, the cells were imaged live after the end of exposure time using a laser scanning confocal microscope (LSM710, Zeiss, Oberkochen Germany).
Assessment of oxidative DNA damage by comet assay
Briefly, 1.5 × 105 cells/well were seeded on 6-well plates 24 h before exposure. Cells were exposed to NM400 for 24 h or 72 h and mean % DNA in tails from 100 comets/gel was determined. To detect oxidative DNA damage, post-lysis incubation with the Fpg enzyme was also carried out. Fpg converts oxidized purines to strand breaks, thus allowing indirect readout of oxidative base lesions.
Measurement of apoptosis
Levels of 35 apoptosis-related proteins were analyzed by employing a protein array (Proteome Profiler™ Human Apoptosis Array) following the protocol from the manufacturer. The array was run only once with independent biological replicates from each exposure because of the nature of the experiment design.
Proteomic analyses
Protein precipitation and identification by LC-MS/MS
Cellular proteins were precipitated by methanol/chloroform/water precipitation. After removing the supernatant, the protein pellet was reconstituted in 50 mM ammonium bicarbonate. Next, proteins were reduced with 1.5 µmol of DTT, and then alkylated using 6 µmol of iodoacetamide. Finally, proteins were digested using 1.25 µg trypsin at 37 °C overnight. Peptides were dried in a vacuum concentrator and then resuspended in 0.1% formic acid.
LC-MS/MS analysis was performed on an EASY-nLC 1000 UPLC system interfaced with an Orbitrap Elite mass spectrometer via a Nanospray Flex ion source. Details are described in the companion Supplementary Materials and methods.
Mass spectrometry data analysis
Raw files from the experiments were inspected with Preview 2.3.5 to determine optimal search criteria (Kil et al. Citation2011). These were plugged in MaxQuant v 1.5.5.1 (Cox and Mann Citation2008), mapping the spectra over Human canonical proteome with isoforms downloaded in May 2016 from Uniprot (Boutet et al. Citation2016). The details are described in the online Supplementary Materials and methods.
For comparative proteomics analysis of MWCNT-exposed and control cells, given the small number of biological replicates, only those protein groups that had 100% identification in at least one group were considered. Thus, we had two sets of differential protein groups where the first set was exclusively found in one group (marked by p value of 0) and another set in which each group differed from the corresponding group in the first set with Student's t-test p value <0.05. False discovery rate was <0.01.
We refrained from inferring rigorous statistical significance analysis. However, we ensured that our protein groups identification themselves were set at the level of FDR <0.01 using target-decoy protein search strategy.
Lipidomic analyses
Lipid extraction and quantification by shotgun mass spectrometry
Cell pellets were homogenized in isopropanol, protein concentrations determined by the BCA assay, and 50 µg of total protein was extracted with MTBE as described (Schuhmann et al. Citation2012). Briefly, 700 µL of 10:3 MTBE/MeOH containing one internal standard for each lipid class was added to the dried homogenate. Mass spectrometry analysis was performed on a Q Exactive instrument and using nanoelectrospray chips with a diameter of 4.1 µm. For FT, MS/MS micro-scans were set to 1, isolation window to 0.8 Da, normalized collision energy to 12.5%, AGC to 5 × 104 and a maximum injection time to 3000 ms. PS was additionally measured for 1.5 min in negative FTMS mode with the same parameters as mentioned above. Lipids were identified by LipidXplorer software (Herzog et al. Citation2012). The detailed procedures are described in the companion Supplementary Materials and methods.
Results
Material characterization
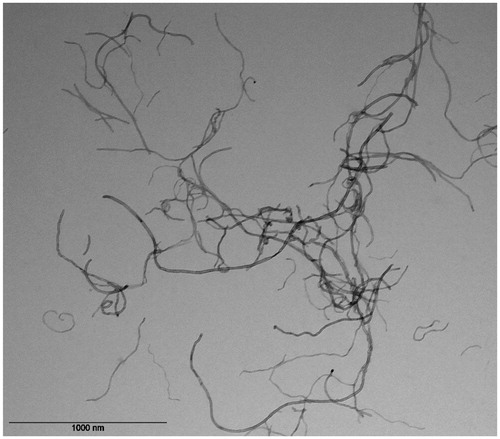
The MWCNT designated as NM400 is a thoroughly characterized JRC reference nanomaterial as described in Materials and methods. We performed further analysis with transmission electron microscopy (TEM) in the exposure medium where the fibers appeared irregular and were mostly entangled and bent (). The observed morphology of the fibers is comparable with the analysis reported by JRC,
Measurement of cytotoxicity, ROS, and oxidative DNA damage
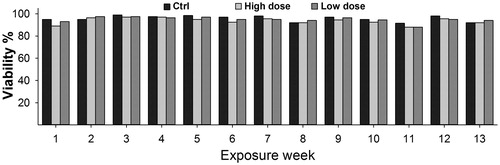
The concentrations 1.92 µg/cm2 and 0.96 µg/cm2 used were not cytotoxic as the viability of NM400-treated cells remained similar to that of the controls throughout the entire exposure period (). However, compared with control cells, low-dose exposed cells showed 30% and high-dose exposed cells showed 25% lower proliferation at the end of exposure by week 13 (Figure S4).
Figure 2. CNT exposure does not affect cell viability. Cells were exposed to 1.92 µg/cm2 (High dose) or 0.92 µg/cm2 (Low dose) of NM400 for 13-weeks. Viability was determined every week using trypan blue assay before sub-culturing cells for new round of exposure. At both concentrations, cell viability of exposed cells remained similar to that of control cells throughout the whole exposure duration. Data represent mean values of biological duplicates for each week.
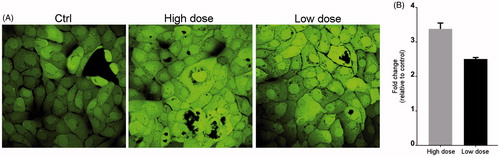
Live cell imaging () showed a marked induction of ROS with both concentrations of NM400. This was further corroborated by the DCF-DA assay which demonstrated a significant generation of ROS ().
Figure 3. CNT exposure increases ROS production. Cells were exposed to NM400 for 72 h, and ROS generation was measured using the DCF-DA assay. (A) Representative LSM image of exposed and control cells. (B) Bar graph showing relative fluorescence per µg protein from two independent experiments each performed with three replicates. Error bars = standard error of mean (SEM); *statistically significant difference relative to control (p < 0.05, paired t-test). High dose:1.92 µg/cm2 and low dose: 0.96 µg/cm2.
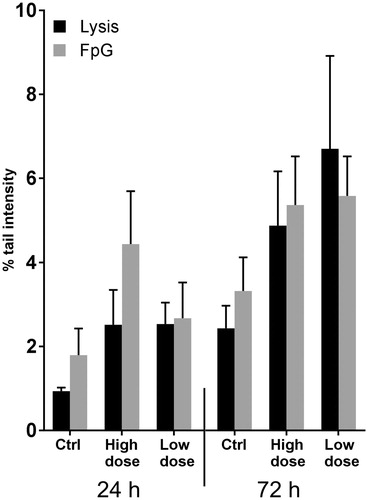
Despite increased ROS levels, comet assay alone or with Fpg showed no significant changes in oxidative DNA damage ().
Figure 4. Comet assay shows no DNA damage after CNT exposure. Oxidative DNA damage after treatment of cells with 1.92 µg/cm2 (high dose) and 0.96 µg/cm2 (low dose) was evaluated using a modified comet assay. No major changes in DNA damage was observed at any time point. Graphs show average of three independent experiments all performed in triplicates (error bars = SEM).
Alteration of apoptotic proteins
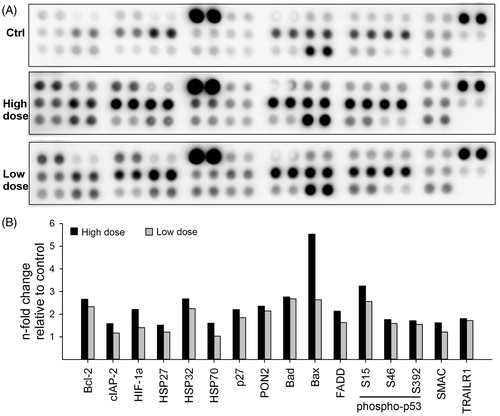
Apoptosis was measured by analyzing both pro- and anti-apoptotic proteins in the HBEC-3KT cells exposed to the MWCNT for 13-weeks, and non-exposed passage-matched control cells (). Quantification of the proteins revealed at least 1.5-fold increased expression, relative to the control, in 16 out of the 35 proteins in either the high or low dose (). BAX was the most upregulated pro-apoptotic protein (∼8-fold in high dose), whereas BCL2 was the most upregulated (∼3-fold in high dose, ∼2.5-fold in low dose) anti-apoptotic protein. In addition, HIF-1α and HSP70 were also expressed at higher levels. Induction of caspase3 cleavage was not observed in the array (spots 9/10 and 11/2 in the upper row of each array represent procaspase 3 and cleaved caspase 3, respectively), indicating that the apoptotic response of the cells was independent of caspase 3 signaling. Thus, the concomitant up-regulation of both pro- and anti-apoptotic proteins apparently balances to avoid apoptosis in the presence of increased ROS.
Figure 5. CNT exposure changes expression of apoptotic proteins. HBEC-3KT cells were exposed to NM400 for 13 weeks, lysed, and 300 µg protein from each condition was used for apoptosis protein array. Because of the nature of experiment, the array was possible to run only once with independent biological replicates for each exposure. (A) representative protein arrays; (B) densitometric analysis of the array spots. Only proteins that changed more than 1.5 fold compared with control are shown. High dose: 1.92 µg/cm2 and low dose: 0.96 µg/cm2.
Comparative proteomics analysis of MWCNT-exposed and control cells
The proteomes of the MWCNT-treated and untreated HBEC-3KT cells were characterized by label-free quantitative mass spectrometry, where more than 5000 proteins were detected across all samples. The complete dataset with the search results has been submitted to PRIDE (identifier PXD005970). For the comparative analysis of exposed and unexposed cells, we first considered proteins that were detected in all three samples. The volcano plots in illustrate the relative abundances of proteins in exposed cells compared with control (). The abundance of proteins within the exposed cells for the high dose compared with the low dose is illustrated in . The plots for high dose versus control and low dose versus control are slightly skewed to the left indicating that a large number of proteins are downregulated. Overall, the number of significantly downregulated proteins was higher than the number of upregulated proteins ().
Figure 6. Volcano plots of all detected and quantified proteins. Plots illustrate the overall pattern of protein expression and their relative abundances in high dose versus Ctrl (A), low dose versus Ctrl (B) or high dose versus low dose (C). The horizontal dotted line in each plot represents p < 0.05. The red dots mark the significantly altered proteins.
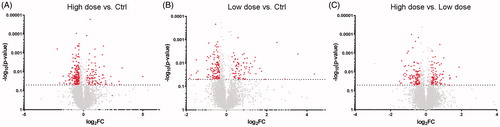
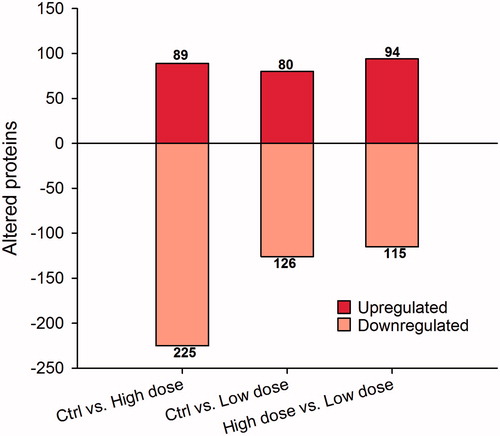
Figure 7. Differentially expressed proteins in CNT-exposed cells. After exposure for 13 weeks to high dose (1.92 µg/cm2) and low dose (0.96 µg/cm2), proteins were extracted from cells and subjected to LC-MS/MS analysis. From >5000 identified proteins, only proteins showing more than 1.2 × fold change compared with control and with p < 0.05 were considered. The actual numbers shown on the top of bars indicate total upregulated or downregulated proteins for each comparison.
To explore and understand the mechanisms of the effects induced after long-term exposure to NM400 (13 weeks), we focused on the differentially regulated proteins that were common to both high dose and low dose compared with control. The list of these proteins is shown in Supplementary Table S1. By setting the fold change cutoff at > |1.5|, 20 proteins were found to be significantly up-regulated (p < 0.05) in high dose/low dose compared with controls (Table S1A), while 17 proteins were significantly down-regulated (p < 0.05) in the CNT-treated cells (Table S1B).
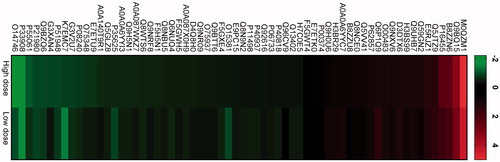
Cellular stress can mediate either very strong up- or down-regulation of proteins to an extent that they are below the detection limit in the untreated- or treated cells, respectively. To include the most significant of these, we chose proteins that were (a) undetectable in controls but detected in at least one of the high or low doses by at least two peptides and a score >5 (Table S1C) and (b) not detected in high dose and low dose, but detected in the controls by the same criteria as in (a) (Table S1D). This added another 20 proteins that were upregulated, and one protein that was downregulated. A heat map of all the differentially regulated proteins is shown in .
Figure 8. Heat-map showing differentially regulated proteins in two exposure conditions high dose and low dose compared with control. First, the different CNT exposure conditions were compared with control. Thereafter, the common proteins were identified from the list of differentially regulated proteins. Figure shows fold change in log2 scale and the protein IDs. Green color indicates proteins that are downregulated in high dose and low dose compared with control, while red color represents upregulated proteins in high dose and low dose compared with control.
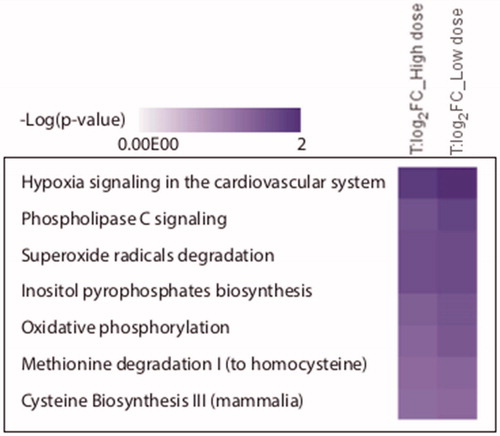
The merged list of differentially expressed proteins was subjected to Ingenuity® pathway analysis (IPA). The most significantly altered pathways in both high dose and low dose are illustrated in . Notably, hypoxia signaling system was the most significantly affected pathway in cells exposed to both high and low doses, whereas superoxide radical degradation and phospholipase c signaling were the second most affected pathways in low dose and high dose, respectively. This corroborates the finding that ROS formation is significantly increased in the CNT-treated cells.
Figure 9. The most affected biological pathways as inferred from the IPA analysis. After comparison of exposed conditions to control, the protein groups with Student’s t-test p values <0.05 and mean absolute log2 LFQ ratio >0.58 were analyzed through the use of QIAGEN’s Ingenuity® Pathway Analysis to determine enriched functional categories. The figure lists biological pathways that are predicted to be regulated by significantly altered proteins from the exposure conditions: high dose and low dose.
Comparative analysis of lipid profiles of MWCNT-exposed and control cells
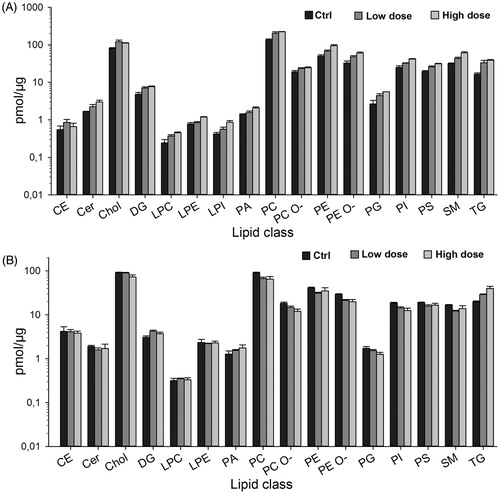
Lipid composition of control and exposed cells was determined by shotgun lipidomics. Interestingly, there was a remarkable increase in total lipids level in exposed compared with control cells (). The total lipids level increased from 430 pmol/µg to 720 pmol/µg and 626 pmol/µg in high dose and low dose, respectively. Correspondingly, there was a concomitant accumulation of most of the lipid classes (). This trend was more pronounced with the highest dose of MWCNT. Triacylglycerol (TG) was highly elevated () at both concentrations followed by sphingomyelin (SM), ceramide (Cer), phosphatidylethanolamine (PE), and cholesterol (Chol). Although cells exposed to the high dose of the MWCNT exhibited an increase in some of the lipid classes compared with the control and the low-dose exposed cells, we refrained from inferring any statistical significance analysis, since the lipidomic study was performed with two biological replicates even though technical duplicates were included during the analysis. The pattern of increase in all lipid classes was intriguing and prompted us to run lipidomic analysis 2 weeks post-exposure. Therefore, exposed cells were reseeded and passaged without MWCNT for 2 weeks, and lipids were extracted and analyzed. Interestingly, the previously observed lipid accumulation as shown in was diminished (). Additionally, the lipid classes such as SM, Cer, PE, and Chol that previously showed high levels tended to slightly decrease (). These data suggest that the continuous and repeated exposure of cells to MWCNT nanomaterial lead to transient accumulation of most of the lipid classes. However, TG remained at similar levels in the high dose indicating a rather stable effect on the cells ().
Figure 10. Lipid classes in exposed versus control cells. Lipids were extracted from the CNT-exposed cells as described in materials and methods, and subjected to shotgun lipidomics. Amount of different lipid classes in cells at the end of 13 weeks of exposure (A) and 2 weeks post-exposure (B) are shown as pmol/µg protein (note the logarithmic scale).
Discussion
Morphologically, NM400 appeared less rigid and less straight than, for example Mitsui-7 that has been classified as a potential carcinogen (Grosse et al. Citation2014). The doses of NM400 used here were not cytotoxic but generated a significant amount of intracellular ROS in the cells which confirms the ability of NM400 to induce ROS as observed in other cell types (Cao et al. Citation2014a, Citation2016). In contrast, we detected no significant DNA strand breaks or oxidized base lesions as it had been reported previously (Kato et al. Citation2013). While ROS are implicated in oxidative DNA damage (Cadet and Wagner Citation2013), the physicochemical properties of CNTs such as size, rigidity, functionalization, and their behavior in exposure media may modulate DNA damaging effects (Wick et al. Citation2007; Nagai et al. Citation2011; Xu et al. Citation2014). Additionally, for non-fibrous nanomaterials, size-dependent levels of DNA damage were observed by the Comet assay, where DNA damage after treatment with ZnO nanoparticles of ≤35 nm was significantly higher than that of NPs of 50–80 nm (Demir et al. Citation2015). The authors also demonstrated that by using Fpg, which recognizes oxidized lesions such as 8-oxoG, only 50–80 nm NPs were able to induce a significant level of oxidative DNA damage. It was also observed that DNA damage was removed in approximately 5 h. Furthermore, exposure to TiO2 NP has been shown to lead to down-regulation of the major DNA repair pathways in human lung cells (Biola-Clier et al. Citation2017). Our proteomics data showed increased expression of the proteins SOSS complex subunit B1 (NABP2) and small ubiquitin-related modifier 1 (SUMO1), both of which play central roles in DNA repair (Huang et al. Citation2009; Jackson and Durocher Citation2013).
Apoptotic responses in MWCNT-exposed cells have been reported (Ravichandran et al. Citation2010; Srivastava et al. Citation2011; Wang et al. Citation2012, Citation2014), but none of the studies have analyzed a large panel of pro- and anti-apoptotic proteins from multiple pathways inducing or antagonizing apoptosis. Since apoptosis is a result of the balance between pro- and anti-apoptotic proteins, arraying them simultaneously is more relevant and realistic. Furthermore, induction of apoptosis could be dose dependent and an advantage of our study is that the cells were exposed to low and non-toxic doses, which may better reflect apoptotic cell death rather than non-specific cell death due to high-dose and toxic overloads. Results from apoptosis protein array revealed a dose dependent, but differential, pattern of protein expression. Notably, exposure to NM400 led to a change in both pro- and anti-apoptotic proteins. Pro-apoptotic proteins Bad, Bax, FADD, and TP53 showed remarkable accumulation, thereby signaling the activation of apoptotic pathways. Proteins contributing to apoptotic resistance, Bcl-2, HSP32, and PON2, also accumulated, albeit to a lesser extent compared with the pro-apoptotic proteins. The overall increase, however, tended to a shift towards pro-apoptotic proteins, indicating induction of apoptotic responses. Functionalized MWCNT has been shown to induce release of cytochrome c and caspase-3/-9 mediated apoptosis in RAW264.7 mouse cells (Wang et al. Citation2012). However, we observed a slight increase of cytochrome c and cleaved caspase-3 only with the highest concentration (data not shown). It is also important to mention that because of the long-term nature of our study, we had limited amount of samples and were thus unable to complement our array data with other apoptosis assays and further studies are required to dissect the role of diverse apoptotic pathways in detail. It should be noted that cells up-regulated PON2 and HIF-1α levels when undergoing oxidative stress, thus, the increased levels of PON2 and HIF-1α in exposed cells could be the result of increased oxidative stress due to high ROS levels.
The proteomic data revealed more than 200 proteins with altered expression. The total number of altered proteins was higher for the high dose concentration of NM400 and the majority were down-regulated. A similar pattern of protein expression has been observed in MWCNT-exposed human keratinocytes (Witzmann and Monteiro-Riviere, Citation2006) but not in human aortic endothelial cells, where the up-regulated proteins dominated (Vidanapathirana et al. Citation2012). Although there was a clear difference between control and CNT exposed cells, however, our data did not show significant dose-dependent alterations since the highest dose always did not result in a significant higher effect in (relative) protein abundance. IPA analysis predicted that the altered proteins are important mediators of different cellular functions, including cell death and survival, cellular assembly and organization, protein folding, and post-translational modifications.
We particularly focused on the altered proteins that displayed similar differential expression at the two concentrations of CNTs. The most up-regulated protein was HNRNPM (∼32- and 21-fold in high dose and low dose, respectively). HNRNPM has been found up-regulated in MWCNT exposed human U937 leukemia cells (Haniu et al. Citation2010). HNRNPM is associated with early spliceosomes and mediates alternative splicing of specific mRNAs (Hovhannisyan and Carstens Citation2007). It was also found to bind internal ribosomal entry sites (IRES) and thereby increasing FGF1 mRNA levels by either increased transcription or mRNA stabilization (Ainaoui et al. Citation2015). It is also involved in TGFβ signaling and promotion of epithelial to mesenchymal transition (Xu et al. Citation2014). It is noteworthy that among the stress response proteins found to be up-regulated in the apoptosis array (), five of them (BCL2, HIF1A, HSP70 P27, and TP53) are among the human proteins of which their mRNAs contain verified IRES (http://iresite.org/IRESite_web.php?page=browse_cellular_transcripts). The up-regulated proteins, NABP2, SUMO1, and MGMT have DNA repair functions. NABP2 is an ssDNA binding protein that activates DNA damage responses (Richard et al. Citation2008). Under oxidative stress, NABP2 is stabilized, and contributes to recruit the OGG1 DNA glycosylase to 8-oxoG lesions and increased repair of 8-oxoG (Paquet et al. Citation2016). The 7–9-fold increased expression of NABP2 in the NM400-treated cells, likely contributes to the relatively small differences observed in the comet intensities observed in the standard- versus the Fpg-mediated comets (). Sumoylation has important functions in the DNA damage and repair responses (Sarangi and Zhao Citation2015). SUMO E1 and E2 ligases constitute essential thiol switches whose reversible oxidation contribute to cell survival during oxidative stress (Stankovic-Valentin et al. Citation2016). MGMT is a DNA repair enzyme that repairs O6-methylguanine, however, any mechanism by which increased the expression of MGMT might contribute to MWCNT genotoxicity, is unknown. The increased MGMT expression might, however, result from ROS-mediated epigenetic activation, since a low ROS-level has been associated with increased MGMT promoter methylation and silencing (Switzeny et al. Citation2012). There was a ∼2-fold up-regulation of AFF4 protein in high dose and low dose. AFF4 has been assigned a role in oxidative stress response. AFF4 is a constituent of the super-elongation complex (SEC), which is activated by HIF1A to alleviate pausing of RNA polymerase II (Luo et al. Citation2012). Under normoxia conditions, many genes harbor transcriptionally engaged, proximally paused RNAPII. Upon oxidative stress, HIF1A recruits AFF4-containing SEC to allow elongation of hypoxia-inducible transcripts (Galbraith et al. Citation2013). Thus, up-regulated expression of AFF4 might play a protective role against MWCNT-induced ROS.
Among the 20 proteins identified exclusively in CNT-treated cells, two additional factors have defined functions in the DNA damage response, likely mediated by their association to modified histones. The chromobox protein homolog 1 (CBX1 and HP1β) is recruited to sites of oxidative DNA damage and DSBs and apparently has important functions in co-ordinating the activities of other DNA repair proteins and to allow access to DNA lesions in densely packed heterochromatin (Bartova et al. Citation2017). Mortality factor 4-like protein 2 (MORF4L2) is part of the NuA4 histone acetyltransferase complex that locally relaxes the chromatin structure to allow DSB repair (Gursoy-Yuzugullu et al. Citation2016).
A number of proteins were downregulated after MWCNT treatment. Most notably, telomerase reverse transcriptase (TERT) was 4.5-fold down-regulated. This is in contrast to the increased TERT activity observed in tumorigenesis (Artandi et al. Citation2002). However, TERT is known to exhibit non-telomeric functions, including protection against DNA damage, oxidative stress, mitochondrial dysfunction, and apoptosis (Iannilli et al. Citation2013; Singhapol et al. Citation2013). Only one additional protein, FNBP1L (also named TOCA1), was exclusively detected in the controls. FNBP1L is involved in membrane invagination and endocytosis (Watson et al. Citation2016) via FNBP1L-induced actin nucleation through CDC42-dependent activation of the N-WASP-WIP/CR16 complex (Ho et al. Citation2004). CDC42 was not found to be significantly differentially expressed in our dataset. However, ARHGAP21 (ARHGAP10), which is a Rho-GAP protein preferentially acting as a negative regulator of CDC42 (Dubois et al. Citation2005), was detected in both of the CNT-treated samples but not in controls. It is thus tempting to speculate whether this might constitute an adaptive mechanism to preclude endocytosis of MWCNTs, which has been shown to be the mechanism of MWCNT uptake in epithelial and mesothelial cells (Maruyama et al. Citation2015). Finally, although not included in the IPA dataset due to lack of identification in one of the low dose samples, HIF1A-inhibitor HIF1AN was found to be nearly 4-fold down-regulated in the high dose concentration in agreement with the increased ROS and the up-regulated HIF1A expression observed in the apoptosis experiments.
Hilton et al. (Citation2017) using mouse alveolar epithelial cells in vitro investigated the effects of pristine (uncoated) MWCNTs and surface coated (functionalized) CNTs with zinc oxide (ZnO) or aluminum oxide (Al2O3) metal oxides. They found distinct proteomic profiles in mTOR/eIF4/p70S6K signaling and Nrf-2 mediated oxidative stress response pathways. The uncoated MWCNT had increased the expression of the interleukin-1 (IL-1) signaling whereas ZnO-coated MWCNT had increased the Nrf-2 mediated signaling, and inhibition of angiogenesis by thrombospondin-1, oxidative phosphorylation, and mitochondrial dysfunction was increased following Al2O3-coated MWCNT exposure. These results show that both pristine and metal oxide-coated CNTs may lead to induction of protein responses related to lung inflammation and fibrosis but metal coatings may have higher and differential protein responses compared with uncoated MWCNT. Even though we did not find significant changes in the pathways reported by Hilton et al. in our present proteomic analysis in human cells; however, we have previously reported in mouse cells that IL-1 signaling may play a role in MWCNT-induced cellular responses (Arnoldussen et al. Citation2015). Furthermore, it was also shown that the CNTs may affect inter-cellular communication and that IL-1 may modulate these effects in mouse cells (Arnoldussen et al. Citation2016). These discrepancies may be due to several factors including differences in physicochemical properties of the CNTs, doses, and exposure times used, and also indicate inter-species differences between human and mouse cells in cellular responses to CNTs.
Accumulation of neutral lipids such as triacylglycerol, diacylglycerol, and cholesterol, has been demonstrated after MWCNT treatment (Tsukahara and Haniu Citation2011; Cao et al. Citation2014a, Citation2014b). However, a thorough characterization of lipid composition after chronic MWCNT-exposure has not been reported. Our results also showed a dose-dependent accumulation of almost all lipid classes after 13-weeks of exposure. The explanation for this global lipid accumulation in exposed cells is not clear. However, the proteomics data revealed that inositol hexakisphosphate kinase 1 (IP6K1) was robustly identified in high dose and low dose, whereas it was below the level of detection in the controls. IP6K1 was reported to enhance cellular fat accumulation by diminishing AMPK-mediated energy expenditure in cells devoid of the uncoupling protein 1 (UCP1) (Zhu et al. Citation2017). It is also possible that increased ROS may contribute to the observed lipid accumulation. This is supported by previous studies showing that HIF-1α and other transcription factors involved in hypoxic signaling lead to lipid accumulation (Boström et al. Citation2006; Semenza Citation2007). Moreover, Cao et al. (Citation2014a) reported accumulation of neutral lipids in NM400 exposed monocyte-derived foam cells, and attributed this to oxidative stress since N-acetyl cysteine ameliorated the lipid accumulation. We also observed that lipid levels in exposed cells returned to similar levels as in controls after 2 weeks, except for TG. This is interesting since TG accumulation has been linked to ROS-mediated PON-2 up-regulation in murine and human macrophages (Rosenblat et al. Citation2012). Our proteomic data also revealed differential expression of several autophagy-related factors involved in the degradation of cellular lipid droplets, most notably at the high-dose MWCNT concentration. These include EHBP1, RAB7B, RAB10, EHD2, and ATG5. Investigation of hepatocellular lipophagy (Li et al. Citation2016) indicates that RAB7 is involved in recruiting cytoplasmic lipid droplets to autophagosomes, whereas a trimeric complex consisting of RAB10, EHBP1, and EHD2 triggered engulfment of the lipid droplets and lipophagy. Interestingly, we found RAB7B, RAB10, and EHD2 to be ∼2.5-, 1.6-, and 1.5-fold downregulated in high dose and could thus potentially contribute to the increased lipid accumulation in the MWCNT-treated cells.
Concerning correlation between low- and high-dose MWCNT exposure and cellular lipid alterations, a possible dose-dependent increase although moderate, in some lipid classes (i.e. Cer, LPE, LPI, PE, PEO, SM, and TG), was observed as shown in , whereas individual lipid species were marginally affected (data not shown). Similarly, some of the proteins showed dose-dependent changes in the MWCNT-exposed cells as shown in but this dose-dependency was not significant and linear.
Finally, it should be noted that a primary aim of this study has been to investigate cytotoxicity, genotoxicity, and potentially carcinogenic effects associated with continuous exposure to multi-walled carbon nanotubes. Ideally, multiple proteomic and lipidomic analyses should have been undertaken during the course of treatment to firmly establish whether the end-point observations were due to gradual alterations of the protein/lipid pools over time and not the result of one acute treatment. However, throughout the entire experimental period, cells were exposed to fresh MWCNT-containing medium after each sub-culturing. Given the biodurability and high persistence of MWCTs, it is thus very likely that the cells were exposed continuously to intracellular MWCNTs. It is also noteworthy that none of the differentially regulated proteins identified represent acute-phase proteins, substantiating that our experimental setup represents a valid model for long-time exposure to MWCNTs.
Conclusions
Cytotoxicity, apoptosis, oxidative stress, and DNA damage were assessed in human lung epithelial cells chronically exposed to occupationally relevant low doses of a well characterized MWCNT. Furthermore, proteomic and lipidomic approaches were applied to characterize and quantify changes in protein and lipid profiles of the cells. The data indicated elevated ROS, altered apoptotic proteins, and significant changes in protein and lipid composition of the CNT-treated cells. These results shed light on (i) the relationship between long-term and low-dose exposure to MWCNTs and changes in the proteomic profile of human lung epithelial cells which is understudied, and (ii) characterization of lipid composition of CNT-exposed human lung cells, which has not been reported previously. The comprehensive proteomic and lipidomic profiling approach presented here provides a preliminary list of proteins and lipids that could be important mediators of CNT-induced toxicity. Nevertheless, the effects of long-term exposure even at low doses appear to be complex, where multiple proteins, lipids, and cellular functions may be altered simultaneously.
S._PHUYAL_ET_AL._supplementary_content.zip
Download Zip (3 MB)Disclosure statement
The authors declare no conflicts of interest.
Additional information
Funding
References
- Ainaoui, N., F. Hantelys, E. Renaud-Gabardos, M. Bunel, F. Lopez, F. Pujol, R. Planes, et al. 2015. “Promoter-Dependent Translation Controlled by p54nrb and hnRNPM During Myoblast Differentiation.” PLoS One 10 (9): e0136466. doi: 10.1371/journal.pone
- Arnoldussen, Y. J., A. Skogstad, V. Skaug, M. Kasem, A. Haugen, N. Benker, S. Weinbruch, et al. 2015. “Involvement of IL-1 Genes in the Cellular Responses to Carbon Nanotube Exposure.” Cytokine 73 (1): 128–137.
- Arnoldussen, Y. J, K. H. Anmarkrud, V.Skaug, R. N. Apte, A. Haugen, and S. Zienolddiny. 2016. “Effects of Carbon Nanotubes on Intercellular Communication and Involvement of IL-1 Genes.” Journal of Cell Communication and Signaling 10 (2): 153–162.
- Artandi, S. E., S. Alson, M. K. Tietze, N. E. Sharpless, S. Ye, R. A. Greenberg, D. H. Castrillon, et al. 2002. “Constitutive Telomerase Expression Promotes Mammary Carcinomas in Aging Mice.” Proceedings of the National Academy of Sciences of the United States of America 99 (12): 8191–8196.
- Atilla-Gokcumen, G. E., E. Muro, J. Relat-Goberna, S. Sasse, A. Bedigian, M. L. Coughlin, Sergi, Garcia-Manyes, et al. 2014. “Dividing Cells Regulate Their Lipid Composition and Localization.” Cell 156 (3): 428–439.
- Bartova, E., B. Malyskova, D. Komurkova, S. Legartova, J. Suchankova, J. Krejci, and S. Kozubek. 2017. “Function of Heterochromatin Protein 1 During DNA Repair.” Protoplasma 254: 1233–1240.
- Biola-Clier, M., D. Beal, S. Caillat, S. Libert, L. Armand, N. Herlin-Boime, S. Sauvaigo, et al. 2017. “Comparison of the DNA Damage Response in BEAS-2B and A549 Cells Exposed to Titanium Dioxide Nanoparticles.” Mutagenesis 32 (1): 161–172.
- Boström, P., B. Magnusson, P.-A. Svensson, O. Wiklund, J. Borén, L. M. S. Carlsson, M. Ståhlman, S. O. Olofsson, and L. M. Hultén. 2006. “Hypoxia Converts Human Macrophages Into Triglyceride-Loaded Foam Cells.” Arteriosclerosis Thrombosis, and Vascular Biology 26 (8): 1871–1876.
- Boutet, E., D. Lieberherr, M. Tognolli, M. Schneider, P. Bansal, A. J. Bridge, S. Poux, L. Bougueleret, and I. Xenarios. 2016. “Uniprotkb/Swiss-prot, the Manually Annotated Section of the Uniprot Knowledgebase: How to use the Entry View.” Methods in Molecular Biology 1374: 23–54.
- Cadet, J., and J. R. Wagner. 2013. “DNA Base Damage by Reactive Oxygen Species, Oxidizing Agents, and UV Radiation.” Cold Spring Harbor Perspectives in Biology 5 (2): a012559.
- Cao, Y., N. R. Jacobsen, P. H. Danielsen, A. G. Lenz, T. Stoeger, S. Loft, H. Wallin, M. Roursgaard, L. Mikkelsen, and P. Møller. 2014a. “Vascular Effects of Multiwalled Carbon Nanotubes in Dyslipidemic ApoE-/- Mice and Cultured Endothelial Cells.” Toxicological Sciences 138: 104–116.
- Cao, Y., M. Roursgaard, P. H. Danielsen, P. Møller, and S. Loft. 2014b. “Carbon Black Nanoparticles Promote Endothelial Activation and Lipid Accumulation in Macrophages Independently of Intracellular ROS Production.” PLoS One 9: e106711.
- Cao, Y., M. Roursgaard, N. R. Jacobsen, P. Møller, and S. Loft. 2016. “Monocyte Adhesion Induced by Multi-walled Carbon Nanotubes and Palmitic Acid in Endothelial Cells and Alveolar–endothelial Co-cultures.” Nanotoxicology 10: 235–244.
- Cox, J., and M. Mann. 2008. “MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-range Mass Accuracies and Proteome-wide Protein Quantification.” Nature Biotechnology 26 (12): 1367–1372.
- De Volder, M. F. L., S. H. Tawfick, R. H. Baughman, and A. J. Hart. 2013. “Carbon Nanotubes: Present and Future Commercial Applications.” Science 6119: 339.
- Demir, E., H. Akça, F. Turna, S. Aksakal, D. Burgucu, B. Kaya, O. Tokgün, et al. 2015. “Genotoxic and Cell-transforming Effects of Titanium Dioxide Nanoparticles.” Environmental Research 136: 300–308.
- Donaldson, K., C. A. Poland, F. A. Murphy, M. Macfarlane, T. Chernova, and A. Schinwald. 2013. “Pulmonary Toxicity of Carbon Nanotubes and Asbestos: Similarities and Differences.” Advanced Drug Delivery Reviews 65 (15): 2078–2086.
- Dubois, T., O. Paléotti, A. A. Mironov, V. Fraisier, T. E. B. Stradal, M. A. De Matteis, M. Franco, et al. 2005. “Golgi-localized GAP for Cdc42 Functions Downstream of ARF1 to Control Arp2/3 Complex and F-actin Dynamics.” Nature Cell Biology 7 (4): 353–364.
- Galbraith, M. D., M. A. Allen, C. L. Bensard, X. Wang, M. K. Schwinn, B. Qin, H. W. Long, et al. 2013. “HIF1A Employs CDK8-mediator to Stimulate RNAPII Elongation in Response to Hypoxia.” Cell 153 (6): 1327–1339.
- Grosse, Y., Loomis, D., Guyton, K. Z., Lauby-Secretan, B., El Ghissassi, F., Bouvard, V., et al. 2014. “Carcinogenicity of Fluoro-edenite, Silicon Carbide Fibres and Whiskers, and Carbon Nanotubes.” Lancet Oncology 15: 1427–1428.
- Gursoy-Yuzugullu, O., N. House, and B. D. Price. 2016. “Patching Broken DNA: Nucleosome Dynamics and the Repair of DNA Breaks.” The Journal of Molecular Biology 428 (9 Pt B): 1846–1860.
- Haniu, H., Y. Matsuda, K. Takeuchi, Y. A. Kim, T. Hayashi, and M. Endo. 2010. “Proteomics-based Safety Evaluation of Multi-walled Carbon Nanotubes.” Toxicology and Applied Pharmacology 242 (3): 256–262.
- Herzog, R., K. Schuhmann, D. Schwudke, J. L. Sampaio, S. R. Bornstein, M. Schroeder, A. Shevchenko, et al. 2012. “LipidXplorer: A Software for Consensual Cross-platform Lipidomics.” PLoS One 7 (1): e29851.
- Hilton, G. M., A. J. Taylor, C. D. Mcclure, G. N. Parsons, J. C. Bonner, and M. S. Bereman. 2015. “Toxicoproteomic Analysis of Pulmonary Carbon Nanotube Exposure Using LC-MS/MS.” Toxicology 329: 80–87.
- Hilton, G. M., A. J. Taylor, S. Hussain, E. C. Dandley, E. H. Griffith, S. Garantziotis, G. N. Parsons, et al. 2017. “Mapping Differential Cellular Protein Response of Mouse Alveolar Epithelial Cells to Multi-walled Carbon Nanotubes as a Function of Atomic Layer Deposition Coating.” Nanotoxicology 11 (3): 313–326.
- Ho, H. Y., R. Rohatgi, A. M. Lebensohn, M. Le, J. Li, S. P. Gygi, and M. W. Kirschner. 2004. “Toca-1 Mediates Cdc42-Dependent Actin Nucleation by Activating the N-WASP-WIP Complex.” Cell 118: 203–216.
- Hovhannisyan, R. H., and R. P. Carstens. 2007. “Heterogeneous Ribonucleoprotein m is a Splicing Regulatory Protein that can Enhance or Silence Splicing of Alternatively Spliced Exons.” The Journal of Biological Chemistry 282 (50): 36265–36274.
- Huang, J., Z. Gong, G. Ghosal, and J. Chen. 2009. “SOSS Complexes Participate in the Maintenance of Genomic Stability.” Molecular Cell 35 (3):384–393.
- Iannilli, F., F. Zalfa, A. Gartner, C. Bagni, C. G. Dotti, F. Di Cunto, 2013. “Cytoplasmic TERT Associates to RNA Granules in Fully Mature Neurons: Role in the Translational Control of the Cell Cycle Inhibitor p15INK4B. PLoS One 8 (6): e66602.
- Iijima, S. 1991. “Helical Microtubules of Graphitic Carbon.” Nature 354 (6348): 56–58.
- Jackson, S. P., and D. Durocher. 2013. “Regulation of DNA Damage Responses by Ubiquitin and SUMO.” Molecular Cell 49 (5): 795–807.
- Jensen, K. A., Y. Kembouche, E. Christiansen, N. R. Jacobsen, H. Wallin, C. Guiot, O. Spalla, and O. Witschger. 2011. “The generic NANOGENOTOX dispersion protocol.” Standard Operation Procedure (SOP) and background documentation Final Protocol for producing suitable manufactured nanomaterial exposure media.
- Jia, X., and F. Wei. 2017. “Advances in Production and Applications of Carbon Nanotubes.” Topics in Current Chemistry (Cham) 375:18.
- Ju, L., G. Zhang, X. Zhang, Z. Jia, X. Gao, Y. Jiang, C. Yan, et al. 2014. “Proteomic Analysis of Cellular Response Induced by Multi-Walled Carbon Nanotubes Exposure in A549 Cells.” PLoS One 9 (1): e84974.
- Kasai, T., Y. Umeda, M. Ohnishi, H. Kondo, T. Takeuchi, S. Aiso, T. Nishizawa, et al. 2015. “Thirteen-week Study of Toxicity of Fiber-like Multi-walled Carbon Nanotubes with Whole-body Inhalation Exposure in Rats.” Nanotoxicology 9 (4): 413–422.
- Kato, T., Y. Totsuka, K. Ishino, Y. Matsumoto, Y. Tada, D. Nakae, S. Goto, , et al. 2013. “Genotoxicity of Multi-walled Carbon Nanotubes in Both In vitro and In vivo Assay Systems.” Nanotoxicology 7 (4): 452–461.
- Kil, Y. J., C. Becker, W. Sandoval, D. Goldberg, and M. Bern. 2011. “Preview: A Program for Surveying Shotgun Proteomics Tandem Mass Spectrometry Data.” Analytical Chemistry 83 (13): 5259–5267.
- Li, Z., R. J. Schulze, S. G. Weller, E. W. Krueger, M. B. Schott, X. Zhang, C. A. Casey, et al. 2016. A Novel Rab10-EHBP1-EHD2 Complex Essential for the Autophagic Engulfment of Lipid Droplets.” Science Advances 2 (12): e1601470.
- Luo, Z., C. Lin, and A. Shilatifard. 2012. “The Super Elongation Complex (SEC) Family in Transcriptional Control.” Nature Reviews Molecular Cell Biology 13 (9): 543–547.
- Maruyama, K., H. Haniu, N. Saito, Y. Matsuda, T. Tsukahara, S. Kobayashi, M. Tanaka, et al. 2015. “Endocytosis of Multiwalled Carbon Nanotubes in Bronchial Epithelial and Mesothelial Cells.” BioMed Research International 2015: 793186.
- Muller, J., F. Huaux, N. Moreau, P. Misson, J. -F. Heilier, M. Delos, M. Arras, et al. 2005. “Respiratory Toxicity of Multi-wall Carbon Nanotubes.” Toxicology and Applied Pharmacology 207 (3): 221–231.
- Muro, E., G. E. Atilla-Gokcumen, and U. S. Eggert. 2014. “Lipids in Cell Biology: How can we Understand them Better? Molecular Biology of the Cell 25 (12): 1819–1823.
- Nagai, H., Y. Okazaki, S. H. Chew, N. Misawa, Y. Yamashita, S. Akatsuka, T. Ishihara, et al. 2011. “Diameter and Rigidity of Multiwalled Carbon Nanotubes are Critical Factors in Mesothelial Injury and Carcinogenesis.” Proceedings of the National Academy of Sciences of the United States of America 108 (9): E1330–E1338.
- Paquet, N., M. N. Adams, N. W. Ashton, C. Touma, R. Gamsjaeger, L. Cubeddu, V. Leong, et al. 2016. “hSSB1 (NABP2/OBFC2B) is Regulated by Oxidative Stress.” Scientific Reports 6: 27446.
- Poland, C. A., R. Duffin, I. Kinloch, A. Maynard, W. A. H. Wallace, A. Seaton, V. Stone, et al. 2008. “Carbon Nanotubes Introduced into the Abdominal Cavity of Mice Show Asbestos-like Pathogenicity in a Pilot Study.” Nature Nanotechnology 3: 423–428.
- Polimeni, M., G. R. Gulino, E. Gazzano, J. Kopecka, A. Marucco, I. Fenoglio, F. Cesano, et al. 2015. “Multi-walled Carbon Nanotubes Directly Induce Epithelial-mesenchymal Transition in Human Bronchial Epithelial Cells via the TGF-β-Mediated Akt/GSK-3β/SNAIL-1 Signalling Pathway.” Particle and Fibre Toxicology 13 (1): 27.
- Ravichandran, P., S. Baluchamy, B. Sadanandan, R. Gopikrishnan, S. Biradar, V. Ramesh, J. C. Hall, et al. 2010. “Multiwalled Carbon Nanotubes Activate NF-κB and AP-1 Signaling Pathways to Induce Apoptosis in Rat Lung Epithelial Cells.” Apoptosis 15 (12): 1507–1516.
- Richard, D. J., E. Bolderson, L. Cubeddu, R. I. M. Wadsworth, K. Savage, G. G. Sharma, M. L. Nicolette, et al. 2008. “Single-stranded DNA-Binding Protein hSSB1 is Critical for Genomic Stability.” Nature 453 (7195): 677–681.
- Rittinghausen, S., A. Hackbarth, O. Creutzenberg, H. Ernst, U. Heinrich, A. Leonhardt, D. Schaudien, et al. 2014. “The Carcinogenic Effect of Various Multi-walled Carbon Nanotubes (MWCNTs) After Intraperitoneal Injection in Rats.” Particle and Fibre Toxicology 11 (1): 59.
- Rosenblat, M., N. Volkova, N. Paland, and M. Aviram. 2012. “Triglyceride Accumulation in Macrophages Upregulates Paraoxonase 2 (PON2) Expression via ROS-Mediated JNK/c-Jun Signaling Pathway Activation.” BioFactors 38 (6): 458–469.
- Sarangi, P., and X. Zhao. 2015. “SUMO-mediated Regulation of DNA Damage Repair and Responses.” Trends in Biochemical Sciences 40 (4): 233–242.
- Sargent, L. M, D. W. Porter, L. M. Staska, A. F. Hubbs, D. T. Lowry, L. Battelli, K. J. Siegrist, et al. 2014. “Promotion of Lung Adenocarcinoma Following Inhalation Exposure to Multi-walled Carbon Nanotubes.” Particle and Fibre Toxicology 11 (1): 3.
- Schuhmann, K., R. Almeida, M. Baumert, R. Herzog, S. R. Bornstein, and A. Shevchenko. 2012. “Shotgun Lipidomics on a LTQ Orbitrap Mass Spectrometer by Successive Switching Between Acquisition Polarity Modes.” Journal of Mass Spectrometry 47 (1): 96–104.
- Semenza, G. L. 2007. “Oxygen-dependent Regulation of Mitochondrial Respiration by Hypoxia-inducible Factor 1. Biochemical Journal 405 (1): 1–9.
- Singhapol, C., D. Pal, R. Czapiewski, M. Porika, G. Nelson, G. C. Saretzki, J. Santos, 2013. “Mitochondrial Telomerase Protects Cancer Cells from Nuclear DNA Damage and Apoptosis.” PLoS One 8 (1): e52989.
- Snyder, R. J., S. Hussain, A. B. Rice, and S. Garantziotis. 2014. “Multiwalled Carbon Nanotubes Induce Altered Morphology and Loss of Barrier Function in Human Bronchial Epithelium at Noncytotoxic Doses.” International Journal of Nanomedicine 9: 4093–4105.
- Srivastava, R. K, A. B. Pant, M. P. Kashyap, V. Kumar, M. Lohani, L. Jonas, Q. Rahman, 2011. “Multi-walled Carbon Nanotubes Induce Oxidative Stress and Apoptosis in Human Lung Cancer Cell Line-A549. Nanotoxicology 5 (2): 195–207.
- Stankovic-Valentin, N., K. Drzewicka, C. Konig, E. Schiebel, and F. Melchior. 2016. “Redox Regulation of SUMO Enzymes is Required for ATM Activity and Survival in Oxidative Stress.” EMBO Journal 35 (12): 1312–1329.
- Switzeny, O. J., E. Müllner, R. K. H. Wagne, H. Brath, E. Aumüller, and A. G. Haslberger. 2012. “Vitamin and Antioxidant Rich Diet Increases MLH1 Promoter DNA Methylation in DMT2 Subjects.” Clinical Epigenetics 4 (1): 19.
- Tsukahara, T., and H. Haniu. 2011. “Nanoparticle-mediated Intracellular Lipid Accumulation During C2C12 Cell Differentiation.” Biochemical and Biophysical Research Communications 406 (4): 558–563.
- Ursini, C. L., D. Cavallo, A. M. Fresegna, A. Ciervo, R. Maiello, G. Buresti, S. Casciardi, S. Bellucci, and S. Iavicoli. 2014. “Differences in Cytotoxic, Genotoxic, and Inflammatory Response of Bronchial and Alveolar Human Lung Epithelial Cells to Pristine and COOH-Functionalized Multiwalled Carbon Nanotubes.” BioMed Research International 2014: 359506.
- Vidanapathirana, A. K., X. Lai, S. C. Hilderbrand, J. E. Pitzer, R. Podila, S J. Sumner, T. R. Fennell, C. J. Wingard, F. A. Witzmann, J. M. Brown, 2012. “Multi-walled Carbon Nanotube Directed Gene and Protein Expression in Cultured Human Aortic Endothelial Cells is Influenced by Suspension Medium.” Toxicology 302 (2–3): 114–122.
- Wang, L., T. A. Stueckle, A. Mishra, R. Derk, T. Meighan, V. Castranova, Y. Rojanasakul, 2014. “Neoplastic-like Transformation Effect of Single-walled and Multi-walled Carbon Nanotubes Compared to Asbestos on Human Lung Small Airway Epithelial Cells.” Nanotoxicology 8 (5): 485–507.
- Wang, X., J. Guo, T. Chen, H. Nie, H. Wang, J. Zang, X. Cui, G. Jia, 2012. “Multi-walled Carbon Nanotubes Induce Apoptosis via Mitochondrial Pathway and Scavenger Receptor.” Toxicology In Vitro 26 (6): 799–806.
- Watson, J. R., H. M. Fox, D. Nietlispach, J. L. Gallop, D. Owen, and H. R. Mott. 2016. “Investigation of the Interaction Between Cdc42 and its Effector TOCA1: Handover of Cdc42 to the Actin Regulator n-wasp is Facilitated by Differential Binding Affinities.” The Journal of Biological Chemistry 291: 13875–13890.
- Wick, P., P. Manser, L. Limbach, U. Dettlaffweglikowska, F. Krumeich, S. Roth, W. Stark, and A. Bruinink. 2007. “The Degree and Kind of Agglomeration Affect Carbon Nanotube Cytotoxicity.” Toxicology Letters 168 (2): 121–131.
- Witzmann, F. A., and N. A. Monteiro-Riviere. 2006. “Multi-walled Carbon Nanotube Exposure Alters Protein Expression in Human Keratinocytes.” Nanomedicine: Nanotechnology, Biology and Medicine 2 (3): 158–168.
- Xu, J., D. B. Alexander, M. Futakuchi, T. Numano, K. Fukamachi, M. Suzui, T. Omori, et al. 2014. “Size- and Shape-dependent Pleural Translocation, Deposition, Fibrogenesis, and Mesothelial Proliferation by Multiwalled Carbon Nanotubes.” Cancer Science 105 (7): 763–769.
- Xu, J., M. Futakuchi, H. Shimizu, D. B. Alexander, K. Yanagihara, K. Fukamachi, M. Suzui, et al. 2012. “Multi-walled Carbon Nanotubes Translocate into the Pleural Cavity and Induce Visceral Mesothelial Proliferation in Rats.” Cancer Science 103 (12): 2045–2050.
- Zhu, Q., S. Ghoshal, R. Tyagi, and A. Chakraborty. 2017. “Global IP6K1 Deletion Enhances Temperature Modulated Energy Expenditure Which Reduces Carbohydrate and Fat Induced Weight Gain.” Molecular Metabolism 16: 73–85.