ABSTRACT
About 15% of couples experience difficulty in conceiving a child, of which half of the cases are thought to be male-related. Asthenozoospermia, or low sperm motility, is one of the frequent types of male infertility. Although energy metabolism is suggested to be central to the etiology of asthenozoospermia, very few attempts have been made to identify its underlying metabolic pathways. Here, we reconstructed SpermNet, the first proteome-scale model of the sperm cell by using whole-proteome data and the mCADRE algorithm. The reconstructed model was then analyzed using the COBRA toolbox. Genes were knocked-out in the model to investigate their effect on ATP production. A total of 78 genes elevated ATP production rate considerably of which most encode components of oxidative phosphorylation, fatty acid oxidation, the Krebs cycle, and members of the solute carrier 25 family. Among them, we identified 11 novel genes which have previously not been associated with sperm cell energy metabolism and may thus be implicated in asthenozoospermia. We further examined the reconstructed model by in silico knock out of currently known asthenozoospermia implicated-genes that were not predicted by our model. The pathways affected by knocking out these genes were also related to energy metabolism, confirming previous findings. Therefore, our model not only predicts the known pathways, it also identifies several non-glycolytic genes for deficient energy metabolism in asthenozoospermia. Finally, this model supports the notion that metabolic pathways besides glycolysis such as oxidative phosphorylation and fatty acid oxidation are essential for sperm energy metabolism and if validated, may form a basis for fertility recovery.
Abbreviations: mCADRE: metabolic context-specificity assessed by deterministic reaction evaluation; ATP: adenosine triphosphate; RNA: ribonucleic acid; FBA: flux balance analysis; FVA: flux variability analysis; DAVID: database for annotation, visualization and integrated discovery; OXPHOS: oxidative phosphorylation; ETC: electron transfer chain; SLC: solute carrier; DLD: dihydrolypoamide dehydrogenase; DLST: dihydrolypoamide S-succinyl transferase; OGDH: oxoglutarate dehydrogenase; CS: citrate synthase; FH: fumarate hydratase; IDH: isocitrate dehydrogenase; SUCLG1: succinate-CoA ligase; SD: succinate dehydrogenase; HADHA: hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase, subunit A; HADHB: hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase, subunit B; PPA2: pyrophosphatase (inorganic) 2; PPi: inorganic phosphate; GALT: galactose-1-phosphate uridylyltransferase
Introduction
Infertility is defined as failure to conceive after having unprotected intercourse for 12 months [Gurunath et al. Citation2011]. It is estimated that about 15% of couples suffer from infertility, with male factors thought to be responsible for about half of these cases [Hwang et al. Citation2011]. Sperm motility, morphology, and concentration are the main parameters affecting semen quality, and are thus routinely evaluated in clinical settings [Amaral et al. Citation2014]. Of these, low sperm motility (asthenozoospermia) is one of the most frequent types of male infertility explaining more than 40% of cases [Liu et al. Citation2015].
Although environmental factors such as unhealthy lifestyle, pollutants, and prolonged duration of sexual abstinence are thought to affect asthenozoospermia, genetic factors are thought to be mainly responsible for the underlying etiology [De Rosa et al. Citation2003; Sharpe Citation2010; Yu et al. Citation2014; Eslamian et al. Citation2015]. Asthenozoospermia has been extensively studied at the molecular level. For instance, comparative proteomic analyses of healthy and asthenozoospermic individuals were used to identify proteins linked to asthenozoospermia [Martínez-Heredia et al. Citation2008; Chan et al. Citation2009; Amaral et al. Citation2013a; Shen et al. Citation2013; Amaral et al. Citation2014]. Other studies have focused on mouse knockout models and have identified novel genes such as Tekt4 and Smcp [Nayernia et al. Citation2002; Roy et al. Citation2007] . In addition, a number of studies have identified genetic polymorphisms associated with asthenozoospermia in multiple genes including Tektin-t, CATSPER1, and eNOS [Zuccarello et al. Citation2008; Visser et al. Citation2011; Song et al. Citation2015]. Despite all these, the underlying etiology of asthenozoospermia has yet to be explicated [Luconi et al. Citation2006; Martínez-Heredia et al. Citation2008; Du et al. Citation2015].
Several structural and functional proteins have been suggested to be responsible for sperm motility [Chan et al. Citation2009; Amaral et al. Citation2013a; Shen et al. Citation2013]. In addition to cytoskeletal and structural proteins in the sperm flagellum [Amaral et al. Citation2013c; Wang et al. Citation2013], metabolic enzymes have also been implicated [Ruiz-Pesini et al. Citation1998; Agarwal and Said Citation2004; Zhao et al. Citation2007]. Although the link between energy production and sperm motility has been explored in a number of studies [Ford Citation2006; Piomboni et al. Citation2012; Amaral et al. Citation2013b], a systematic study of metabolic genetic deficiencies pertaining to asthenozoospermia has not been undertaken [Amaral et al. Citation2014].
Analysis of metabolic networks to understand disease etiology
Constraint-based reconstruction and analyses of metabolic networks have shown a promising potential in modeling biochemical properties of cells [O’Brien et al. Citation2015]. To explore the complexity of cellular metabolism and to improve our understanding on the genotype-phenotype relationship, modeling and analysis of the omics-scale metabolic network of cells is of great importance [Mardinoglu and Nielsen Citation2012]. A genome-scale metabolic network model is a large system of chemical reactions and metabolites connected to each other by gene-protein-reaction relationships. In constraint-based modeling of cellular metabolism, based on an omics-scale metabolic model, metabolic fluxes which are required for biochemical functions are analyzed. Therefore, dysfunction of biochemical pathways in disease can be modeled by constraining the activity of certain reaction(s) in silico and determining the consequences [Ryu et al. Citation2015; Zhang and Hua Citation2015].
The first genome-scale human metabolic network, Recon1, was reconstructed in 2007 [Duarte et al. Citation2007], with its update, Recon2, released six years later [Thiele et al. Citation2013]. Cell- and tissue-specific metabolic network models have also been generated in a top-down approach by pruning these genome-scale models using proteome and transcriptome data obtained from that tissue [Jerby et al. Citation2010; Lewis et al. Citation2010; Fouladiha et al. Citation2015]. In addition, metabolic network models have been reconstructed to explore the underlying etiology of different tissue-specific disorders [Rezola et al. Citation2014; Salazar et al. Citation2016; Sohrabi-Jahromi et al. Citation2016], resulting in the identification of drug targets and biomarkers [Shlomi et al. Citation2009; Frezza et al. Citation2011; Mardinoglu et al. Citation2014]. Despite the significance of sperm cell metabolism in asthenozoospermia, to the best of our knowledge, no metabolic network model has been reconstructed for this cell. In this study, we reconstruct the first proteome-scale metabolic model of the sperm cell (). The reconstructed model was then used to identify genes whose dysfunction can affect sperm motility through reduced ATP production. Our model successfully predicted several genes with known roles in asthenozoospermia (see ). Furthermore, the model identified 11 non-glycolytic genes that affect energy production efficiency. Mutations affecting the structure or expression of encoded enzymes may thus result in asthenozoospermia.
Figure 1. Model reconstruction procedure and findings. We first collected the sperm-specific proteomic data from databases and literature. Then, after converting the protein data to the corresponding gene data, we used them for reconstructing the sperm-specific metabolic network model. Finally, we used the reconstructed model for analyzing energy metabolism in healthy sperm and in asthenozoospermia. After that, under single knockout analysis, our model predicted several genes that may play a role in asthenozoospermia.
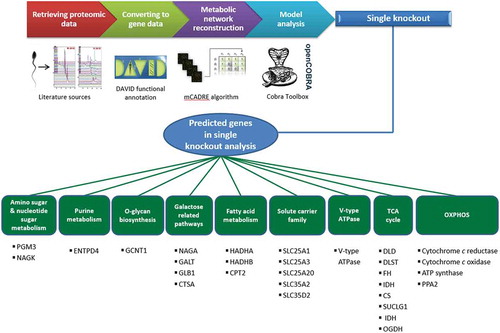
Figure 2. From the total of 29 predicted essential gene sets, 18 gene sets (62%) were previously reported in the literature to be linked to ATP production, and thus, to asthenozoospermia. To the best of our knowledge, the other 11 gene sets (38%) have not previously reported to be linked to asthenozoospermia. The accompanying table shows a list of these genes.
Results and discussion
Proteome-scale reconstruction of the metabolic network in sperm cell
Some studies in recent years provide evidence on the importance of the RNA pool in sperm cells [Johnson et al. Citation2011; Montjean et al. Citation2012], however, the mature spermatozoon is believed to be transcriptionally silent due to its chromatin undergoing condensation during spermatogenesis [Goodrich et al. Citation2013; Jodar et al. Citation2016]. Because of this, we employed the sperm proteomic data for model reconstruction. The model had been manually curated in order to have all essential reactions observed in sperm cell. The reconstructed proteome-scale metabolic network model of the sperm cell, namely SpermNet, is presented in SBML format in Supplementary file S1. This model can be read and analyzed by standard systems biology tools like the COBRA toolbox. SpermNet consists of 2,968 reactions, 2,034 metabolites and 1,242 genes, and is publicly available for those wishing to investigate different aspects of sperm cell metabolism and its related diseases.
In silico gene knockout and model validation
In order to validate the reconstructed model, we investigated its properties using flux balance analysis (FBA). The goal of this analysis was to predict those energy metabolism-related genes whose knockout changes the ATP production rate significantly (i.e., >10%). For this purpose, we knocked out all model genes one by one and explored their knockout effects on ATP production rate of the model. This analysis resulted in the identification of 78 genes listed in Supplementary file S2. Genes which encoded fully coupled reactions [Larhlimi et al. Citation2012] were grouped as a single gene set. Consequently, we obtained 29 independent gene sets in total. Of these, 18 had been previously reported to be linked to ATP production. The model therefore predicted 11 novel genes whose knockout decreased the ATP production rate. Furthermore, we independently analyzed each gene set to understand its role in ATP production in the sperm cell to better understand the underlying pathways affected in asthenozoospermia.
Prediction of down-regulated pathways
To evaluate the effects of each gene knockout on the pathways in our model, we in silico knocked out genes identified by FBA (N=78) one by one and explored their effect on model pathways using flux variability analysis (FVA). Especially, we were interested in high confidence decreased fluxes () because they are likely to be linked to the down-regulated pathways in ATP production, and hence, energy metabolism. We analyzed the high confidence decreased reactions with the database for annotation, visualization, and integrated discovery (DAVID) functional annotation tool [Huang et al. Citation2009]. Interestingly, down-regulated pathways predicted by the model were highly correlated to the experimentally known function of that certain gene. The main down-regulated pathways were ATP metabolism, oxidative phosphorylation, O-glycan metabolism, and fatty acid and lipid metabolism. The full list of down-regulated pathways after each gene knockout is available in Supplementary file S3. The role of each pathway in ATP production and asthenozoospermia is discussed below.
Figure 3. The four reaction categories which show flux decrease or increase in the knockout model based on FVA [Hadi and Marashi Citation2014]. X-axis represents total reactions possible flux ranges. The solid color lines represent the flux of reaction in normal sperm cell model, (ranges from c to d). The dashed arrows indicate the flux of reaction in knockout model (ranges from a to b). If this means a high confidence decrease, however if
it means a high confidence increase. If
, it will result in a decrease, while if
it will result in an increase.
![Figure 3. The four reaction categories which show flux decrease or increase in the knockout model based on FVA [Hadi and Marashi Citation2014]. X-axis represents total reactions possible flux ranges. The solid color lines represent the flux of reaction in normal sperm cell model, (ranges from c to d). The dashed arrows indicate the flux of reaction in knockout model (ranges from a to b). If this means a high confidence decrease, however if it means a high confidence increase. If , it will result in a decrease, while if it will result in an increase.](/cms/asset/b0178c11-7afc-468c-a162-22aff1aec437/iaan_a_1263367_f0003_oc.jpg)
Oxidative phosphorylation
Oxidative phosphorylation (OXPHOS) is one of the most important pathways in all cells. There has been a long debate about the role of OXPHOS in sperm cells. Originally, it was believed that glycolysis was the primary source of ATP production in sperm cells and OXPHOS plays a negligible role in providing energy for this cell type [Storey Citation2008]. However, this idea has been disputed by several studies suggesting an important role for OXPHOS in sperm motility [Marchetti et al. Citation2002; Amaral and Ramalho‐Santos Citation2010; Sousa et al. Citation2011]. The emerging idea is that several non-glycolytic pathways are required to function in the sperm cell to produce sufficient ATP [Amaral et al. Citation2013a; Amaral et al. Citation2013c]. In support of the OXPHOS role in sperm motility, it has been observed that inhibition of different compounds of the electron transfer chain (ETC), despite the availability of glucose, leads to a drop in motility [Ruiz-Pesini et al. Citation2000; John et al. Citation2005]. In support of previous observations, our model predicted genes related to OXPHOS whose knockout reduces the ATP production rate in sperm. These genes can be classified into three different groups of cytochrome c reductase (complex III), cytochrome c oxidase (complex IV), and ATP synthase (complex V) related genes.
The first group predicted by the model consisted of 10 cytochrome c reductase related genes. This protein complex, which is involved in the electron transport chain of mitochondria, catalyzes the oxidation of ubiquinol and the reduction of cytochrome c. This oxidation/reduction reaction results in a net transfer of proton across the membrane which adds to the proton gradient. As a member of ETC, it is not surprising that problems in complex III leads to deficiency in ATP production and impaired sperm motility [Ruiz-Pesini et al. Citation1998]. Moreover, an inhibitor of complex III, antimycin A, is reported to decrease sperm motility [Ford and Harrison Citation1981; Ruiz-Pesini et al. Citation2000]. Interestingly, several studies have shown that treatment of asthenozoospermic patients with coenzyme Q10 (ubiquinol) results in improvement of sperm motility [Balercia et al. Citation2009; Safarinejad et al. Citation2012] which might be as a result of higher activity in complex III.
The second group is the cytochrome c oxidase or complex IV family. Cytochrome c oxidase is a large transmembrane protein complex that catalyzes the final step of the electron transfer chain in eukaryotic mitochondria [Li et al. Citation2006]. This complex is suggested to be a major regulatory site for oxidative phosphorylation [Kadenbach et al. Citation2000]. The main function of this enzyme is catalyzing the transfer of electrons to oxygen to produce water. Transfer of electrons to oxygen is coupled to the translocation of protons and is necessary for ATP synthesis. In line with the model predictions, several studies have shown that levels of different protein members of this family are lower in asthenozoospermic samples than those with normal motility [Ruiz-Pesini et al. Citation1998; Ruiz-Pesini et al. Citation2000; Martínez-Heredia et al. Citation2008]. In addition, it has been observed that inhibition of complex IV results in a dramatic decrease in sperm motility [Brown Citation1995; Pascual et al. Citation1996].
The third group includes ATP synthase related genes. ATP synthase is the last part of the OXPHOS pathway and is responsible for the most important part of this pathway, i.e., generation of ATP. This enzyme uses the energy stored in the proton gradient across the membrane to drive the synthesis of ATP. Different studies have explored the impact of deficiency in ATP synthase genes on sperm motility [Ruiz‐Pesini et al. Citation2007; Piomboni et al. Citation2012]. Moreover, oligomycin, a drug that directly blocks ATP synthesis by mitochondria, is shown to negatively influence sperm motility [Dreanno et al. Citation1999].
Solute carrier family 25
The solute carrier family includes a group of proteins responsible for transporting different metabolites across the mitochondrial membrane [Palmieri Citation2013]. These proteins are reported to be essential for efficient synthesis of ATP in mitochondria [Piomboni et al. Citation2012]. Interestingly, three members of the solute carrier (SLC)25 family, namely SLC25A1, SLC25A3, and SLC25A20 were among the genes predicted to be significant by the model. SLC25A1 is a citrate carrier, which promotes efflux of tricarboxylic citrate to the cytoplasm in exchange for dicarboxylic cytosolic malate [Palmieri and Pierri Citation2010; Sun et al. Citation2010]. To the best of our knowledge, no direct evidence is reported in the literature to support the role of this protein in sperm motility. However, since it is a mitochondrial transporter, it is reasonable to suggest its inactivity may lead to asthenozoospermia, especially that inhibition of this protein is shown to reduce the fluxes of several pathways, including sperm hyperactivated motility related pathways [Cappello et al. Citation2012].
The second protein in this family, SLC25A3, is a phosphate transporter. This protein facilitates the transport of phosphate into mitochondria either by proton co-transport or in exchange for hydroxyl ions. This protein is known to play a fundamental role in ATP synthesis in mitochondria [Mayr et al. Citation2007]. A recent study has reported that the transcript of SLC25A3 is differentially regulated in asthenozoospermia in comparison with the normal control group, suggesting a possible role for this protein in asthenozoospermia [Bansal et al. Citation2015].
SLC25A20 is a carnitine carrier. This protein transports acyl-carnitines into the mitochondrial matrix for their oxidation by the mitochondrial fatty acid-oxidation pathway [Fukushima et al. Citation2013]. It works in conjunction with carnitine palmitoyl transferase II, which oxidizes long-chain fatty acids in mitochondria. Interestingly, our model predicted both genes to have a role in ATP production, and hence, in sperm motility. Carnitine is known to have an important role in sperm motility. Insufficient carnitine in sperm is shown to result in severe motility problems [Tanphaichitr Citation1976; Hinton et al. Citation1979; Agarwal and Said Citation2004]. Interestingly, it has been observed that in asthenozoospermic samples, the ratio of acyl-carnitine to carnitine is greatly reduced, suggesting an improper activity of carnitine transferase [Golan et al. Citation1984; Bartellini et al. Citation1986].
Citric acid cycle
Citric acid cycle (or Krebs cycle) is a chemical pathway used by all aerobic organisms to generate energy from oxidation of acetyl-CoA. In eukaryotic cells, this pathway occurs in mitochondria and is closely related to OXPHOS. The relationship between the Krebs cycle and asthenozoospermia has been investigated [Zhao et al. Citation2007; Martínez-Heredia et al. Citation2008], which showed that Krebs related gene deficiencies are linked to asthenozoospermia. In the present work, our model predicted a number of Krebs cycle genes involved in ATP production of sperm cells, and thus, in asthenozoospermia. Dihydrolypoamide dehydrogenase (DLD), dihydrolypoamide S-succinyl transferase (DLST), oxoglutarate dehydrogenase (OGDH), citrate synthase (CS), fumarate hydratase (FH), isocitrate dehydrogenase (IDH), succinate-CoA ligase (SUCLG1), and succinate dehydrogenase (SD) are the genes predicted by SpermNet to influence ATP production in sperm cells.
DLD, DLST, and OGDH are three enzymatic subunits which work together to form the oxoglutarate dehydrogenase complex (α-ketoglutarate dehydrogenase). This complex converts oxoglutarate to succinyl-CoA. Increase in DLD precursor, which is a sign of decrease in mature DLD, has been observed in different asthenozoospermic patients [Martínez-Heredia et al. Citation2008; Ashrafzadeh et al. Citation2013]. Decrease in OGDH has also been linked with reduced sperm motility, consistent with the predictions of SpermNet [Agarwal et al. Citation2016]. Amaral et al. [Citation2014] reported a lower DLST enzyme ratio in asthenozoospermic samples compared with control samples. SUCLG1, CS, FH, SD, and IDH are important enzymes of the Krebs cycle and deficiency in each of them have been shown to lead to impaired sperm motility [Ruiz-Pesini et al. Citation1998; Ruiz-Pesini et al. Citation2000; Zhao et al. Citation2007; Martínez-Heredia et al. Citation2008; Tomar et al. Citation2010; Tomar et al. Citation2012; Amaral et al. Citation2014]. It should be emphasized that the above-mentioned alterations are genetically heterogeneous and therefore, different patients may have different genetic deficiencies leading to asthenozoospermia.
Fatty acid oxidation
Fatty acid metabolism plays an important role in energy production of cells through the beta-oxidation of fatty acids [Costa et al. Citation1994]. In one study, inhibition of fatty acid oxidation using etomoxir was shown to result in a decrease in sperm motility, indicating the importance of beta-oxidation in energy production of the sperm cell [Amaral et al. Citation2013c]. SpermNet predicted two genes in the fatty acid beta-oxidation pathway to have significant roles in ATP production and thus related to asthenozoospermia. Hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase, subunit A (HADHA) and hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase, subunit B (HADHB) are two subunits of the mitochondrial trifunctional enzyme, which catalyzes three out of four steps of beta-oxidation [Kamijo et al. Citation1994]. Proteomic analysis of human sperm cells has shown that decreased levels of HADHA in asthenozoospermic patients, supporting the relevance of our model prediction [Amaral et al. Citation2014]. This prediction is also in line with the new emerging idea that pathways other than glycolysis and OXPHOS participate in energy production in the sperm cell [Amaral et al. Citation2013c].
Pyrophosphatase
Another protein predicted to be linked with ATP production by SpermNet is pyrophosphatase (inorganic) 2 (PPA2). This enzyme resides on the mitochondrial membrane and is involved in producing inorganic phosphate (PPi) through hydrolyzing pyrophosphate. PPi is a stable, easy-to-store, high energy compound, which is able to substitute for ATP under certain conditions in glycolysis-related reactions such as attenuated respiration [Chi and Kemp Citation2000]. Consistent with the prediction of SpermNet, Yi et al. [Citation2012] showed that PPi plays an important role in sperm motility and impaired PPA2 leads to decreased motility.
V-type ATPase family
Another group of genes predicted by our model is the V-type ATPase family. They are mainly found in the apical membrane of cells that line the epididymis and vas deferens in the male reproductive system, controlling pH of the sperm environment [Nishi and Forgac Citation2002]. The sperm cell needs an acidified environment for efficient motility and viability [Forgac Citation2007]. Interestingly, the V-type ATPase subunit E1, predicted by SpermNet to affect ATP production significantly, is only found in sperm cells and in the acrosome part of spermatids and mature sperms, thus suggesting a dual role in energy metabolism and acidification of the acrosome [Sun-Wada et al. Citation2002]. Furthermore, ATPase subunit A2 has been studied in another work, which showed that lower amounts of this protein will lead to decreased sperm motility [Ota et al. Citation2013].
Double gene knockout in silico simulations
We also in silico knocked out genes in pairs to explore the double gene-knockout effects. For this purpose, first, we removed all genes that have been found in the single knockout simulation (N=78) from our gene list. We then knocked out all remaining genes in pairs and then selected those pairs which collectively reduced the ATP hydrolysis rate more than 10%. A total of 157 gene pairs were found (the complete list of gene pairs is available in Supplementary file S4). After consolidating the fully coupled genes together, 18 sets of gene pairs were identified. Main pairs were involved in OXPHOS and fatty acid beta oxidation complexes, which further confirm the role of these pathways in sperm energy metabolism and motility. Simultaneous knockout of electron transfer flavoprotein, which participates in catalyzing the initial step of the mitochondrial fatty acid beta-oxidation, with the mitochondrial respiratory chain complex I and II proteins are also among the pairs of genes which reduces the ATP hydrolysis rate. Genes related to solute carrier family 25 and the Krebs cycle were also among the identified pairs, both of which were discussed above.
Novel predicted genes as potential biomarkers
SpermNet predicted 11 novel genes which significantly affect ATP production and may thus be associated with asthenozoospermia (see ). To the best of our knowledge, none of these genes have been reported to be mutated or associated with asthenozoospermic patients. These genes may therefore become targets of future experiments aimed to identify unknown causes of asthenozoospermia and if validated, may potentially be used as biomarkers.
Table 1. Novel genes predicted by the SpermNet model to play a role in asthenozoospermia.
Interestingly, galactose-1-phosphate uridylyltransferase (GALT) which catalyzes the second step of the Leloir pathway of galactose metabolism was identified. It has previously been observed that this pathway can be active in the sperm cell and play a role in its energy metabolism [Amaral et al. Citation2013c], although GALT has not been specifically linked to asthenozoospermia. The other ten novel genes predicted by SpermNet are involved in nucleotide interconversion, glucosamine metabolism, and keratan sulfate degradation. Given the function of these genes, it is interesting to speculate that spermatozoa deposited in the female reproductive tract find substrates for these gene-products during their voyage to the oocyte. If validated, this provides support for an underlying gene-environment interaction responsible for efficient sperm motility. These genes therefore need to be examined further to establish their association with asthenozoospermia.
SpermNet, however, did not predict genes related to glycolysis. These results would have been surprising if it were not for those of Amaral et al. [Citation2014] which, based on proteomic data generated from asthenozoospermic patients, showed that expression of glycolytic genes are not significantly altered. This finding contributes to the open discussion about the role of glycolysis and other pathways in sperm energy metabolism. It was initially believed that sperm cell energy is predominantly provided through glycolysis, however, this hypothesis was later challenged by several studies showing that OXPHOS and other pathways also play roles in sperm energy metabolism [Ford Citation2006]. Ford showed that in mouse glycolytic gene knockout sperms experienced a drop in sperm motility [Ford Citation2006]. However, incubation of the exact same cells in glucose free medium lead to proper sperm motility, indicating that glucose is not the only energy source for these cells [Ford Citation2006]. Moreover, independent studies have shown that incubation of human sperm with different ETC complex inhibitors in the presence of glucose will still lead to a rapid drop in sperm motility, suggesting that dysfunction in OXPHOS will also lead to asthenozoospermia even when glycolysis is fully functioning [Ruiz-Pesini et al. Citation2000; John et al. Citation2005].
Based on the findings of Amaral et al. [Citation2014] and SpermNet predictions, we suggest that glycolysis is not the main energy source for sperm motility and correct functioning of pathways like OXPHOS and fatty acid oxidation play a significant role. This suggests that although glycolysis plays a significant role in energy metabolism, sperm cell genes that encode components of non-glycolytic pathways have important roles in asthenozoospermia.
False negatives of SpermNet in predicting asthenozoospermia-implicated genes
Although SpermNet predicted 78 genes with a significant role in ATP production and thus potentially implicated in asthenozoospermia, it failed to predict a number of asthenozoospermia-implicated metabolic genes (see ). This failure is probably due to an incomplete reconstructed model as the generic human model and the proteomic dataset used does not cover all proteins encoded in the human genome. Furthermore, reconstruction algorithms are not error-free and some genes and pathways are likely not represented in the model.
Table 2. List of genes that are known to be linked to asthenozoospermia but have not been predicted to influence ATP production by the SpermNet model.
In order to investigate the knockout effect of these genes on SpermNet, we undertook FBA and FVA analysis. After removing each gene, we detected all activated and inactivated reactions and compiled them separately. This procedure resulted in 307 inactivated and 287 activated genes. The DAVID functional annotation tool identified multiple pathways silenced by deleting these genes based on their overall enrichment score, which is a modified p-value. In other words, DAVID tries to rank the biological significance of gene groups based on their overall p-value scores of all enriched annotation terms. shows the top 10 enriched pathways based on the inactivated genes. Interestingly, most of the silenced pathways were related to energy metabolism, which suggests that SpermNet is capable of predicting their knockout effects correctly. Notably, among these enriched pathways, peroxisome-related pathways are also present, which is consistent with findings of a recent study indicating that these pathways are active in sperm cells and play a role in its metabolism [Amaral et al. Citation2013c].
Figure 4. Enriched pathways based on inactivated or activated genes in gene knockout simulations. (A) Enriched pathways based on 307 inactivated genes as a result of deleting asthenozoospermia-implicated metabolic genes not predicted by SpermNet. (B) Enriched pathways based on 287 activated genes as a result of deleting asthenozoospermia-implicated metabolic genes not predicted by SpermNet.
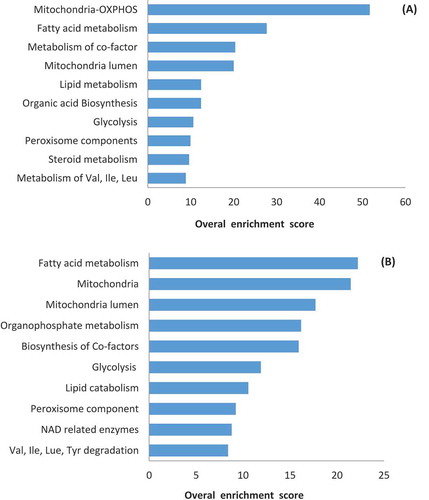
The top 10 enriched pathways based on activated genes are shown in . The underlying basis for gene activation, after deleting asthenozoospermia-implicated metabolic genes from SpermNet, lies with the objective function of the model. Since its aim is to maximize the rate of ATP production, the model achieves this objective by activating other compensatory reactions, hence, up-regulating previously silent genes.
Future study
The first proteome-scale constraint-based metabolic network model of an asthenozoospermia sperm cell was constructed. The model was validated by comparing the predicted genes with asthenozoospermia-implicated genes in the literature, based mainly on mouse knockout models and inhibition assays. However, we believe that these findings have to be validated with in vitro experiments. For instance, knockout mouse models of their orthologs may be used to experimentally validate their functional effect. Mutational analysis of a large cohort of asthenozoospermic patients may also provide evidence for their association.
Additional confirmation can be obtained through drug targeting and then exploring their inhibitory effects on sperm energy metabolism and motility in vitro. This has been done for some genes predicted by SpermNet previously, such as antimycin A which inhibits complex III of ETC and oligomycin which blocks mitochondrial ATP synthase [Dreanno et al. Citation1999; Ruiz-Pesini et al. Citation2000]. For the novel predicted genes, we identified all possible drugs that may target these genes (for a complete list of drugs see Supplementary file S5). Notably, gemcitabine is a drug that inhibits the function of CMPK and can thus be used to analyze its effect on sperm motility [Vernejoul et al. Citation2006]. Further studies analyzing the interaction of potential drugs with these novel genes may have the potential to recover fertility in asthenozoospermic patients.
SpermNet is capable of predicting several related genes to asthenozoospermia and it confirms the emerging idea that several non-glycolytic pathways are implicated in asthenozoospermia and should be considered when analyzing asthenozoospermic patients. The findings presented above suggest that mitochondria and its related pathways (such as OXPHOS) play a significant role in energy metabolism of the sperm cell and any potential deficiency would likely lead to impaired sperm motility.
Materials and methods
Sperm-specific metabolic network reconstruction
In the present study, we used an updated version of Recon2 as the generic template model [Swainston et al. Citation2016]. This model includes 1,713 genes, 5,391 metabolites, and 7,852 reactions. To generate a sperm cell-specific model, proteomic data pertaining to the sperm cell were collected from two sources [Amaral et al. Citation2013a; Wang et al. Citation2013] and meta-analyzed. The consolidated protein list was then converted to a list of Entrez Gene IDs using DAVID [Huang et al. Citation2009]. For any gene being shared in both gene sets, a score of 1 was assigned in the expression array, denoting this gene as active in the sperm-specific metabolic network model. All other genes in the generic model absent in the converted gene set were assigned a score of 0. Among the different algorithms developed for tissue- or cell-specific model reconstruction, we used the metabolic context-specificity assessed by deterministic reaction evaluation (mCADRE) algorithm [Wang et al. Citation2012] for our model reconstruction due to its ability in considering network topology, reducing computational burden, and its known superiority in accurately inferring reactions. This algorithm uses a high-confidence set of core reactions from the human generic template model (e.g., Recon1, Recon2, etc.) based on tissue-specific expression evidence. Other non-core reactions are ranked according to their expression evidence and their link to other network reactions, and then removed in the inverse order of this ranking. Each reaction removal is checked by the consequent flux capacity of core reactions and a universal test of metabolic functionality to see whether that removal should be accepted or rejected [Wang et al. Citation2012]. The reconstructed model was then analyzed using multiple methods in the COBRA Toolbox [Schellenberger et al. Citation2011], such as FBA [Orth et al. Citation2010] and FVA [Mahadevan and Schilling Citation2003].
Flux balance analysis (FBA)
Metabolic network reactions can be represented as an m×n stoichiometric matrix S, in which each column corresponds to a reaction and each row represents a specific metabolite. Flux through all network reactions is represented by vector v. The main assumption in the network analysis is that the system is working on (quasi-)steady state conditions:
Each reaction in the model is either reversible or irreversible. This classification is achieved by restricting each reaction flux with thermodynamic constraints (vimin = 0 for any irreversible reaction i). Furthermore, lower- and upper-bounds might be known for other reactions. All such constraints can be summarized in the “capacity” constraint
where vmin and vmax are the vectors of lower- and upper-bounds of reactions, respectively.
FBA is a mathematical optimization approach which relies on constraints (1) and (2) to gain insight about the reaction fluxes that maximize a biological objective in the model [Orth et al. Citation2010]. Here, we used FBA to explore the contribution of reactions to the maximization of the objective function, adenosine triphosphate (ATP) hydrolysis reaction (ATPasel). ATPasel works as an objective function for maximizing the ATP production, because in order to have the maximum ATP hydrolysis, the model first needs to produce maximum ATP. In this model, 994 reactions should carry flux in order to maximize the objective function.
Knockout simulations
Using constraint-based modeling, one can predict the effect of knocking out any gene in the model on the optimal value of the objective function [Bordbar et al. Citation2014]. Here, we simulated a gene knockout for every single gene in the model and studied these knockout effects on ATP production. This was implemented by the function “SingleGeneDeletion” in the COBRA toolbox [Schellenberger et al. Citation2011]. This function simulates the gene knockout by changing the gene associated reaction bounds to zero, thus inhibiting the flux of those reactions. Furthermore, in every knockout stage, FBA is used to evaluate the amount of ATP hydrolysis in the model by using ATPasel as the objective function. One of the important outputs of this function is grRatio, which is defined as follows:
We decided to have a cutoff of 0.9 for grRatio, which means that we selected any gene whose knockout caused the model to have an ATP hydrolysis rate of less than 90% of that in the normal model (see Supplementary file S6).
A comprehensive list of genes thought to be responsible for asthenozoospermia through deficient energy metabolism was gathered from the literature. The metabolic consequences of gene knockout was simulated for each associated gene using our proteome-scale model. In specific, by using FBA and by setting the flux of ATPasel as the objective function, we identified those reactions that show flux changes in comparison with the fluxes of the normal sperm model. The reactions were then classified into two groups of those which show up-regulation or activation in comparison with the normal model fluxes and those which show the opposite. Reactions belonging to each category had been consolidated separately and then subjected to DAVID functional annotation clustering by DAVID [Huang et al. Citation2009], and ten of the most enriched clusters were selected for further analysis.
Flux variability analysis (FVA)
FVA is a tool for exploring alternative optimal solutions under certain thermodynamic and stoichiometric constraints [Bordbar et al. Citation2014]. It uses FBA-like linear programming problems to find the minimum and maximum flux of each reaction through the network while maintaining the objective function of the network at its optimal value [Mahadevan and Schilling Citation2003].
To evaluate the effect of knocking out each associated gene on the ATP hydrolysis rate, the gene was removed from the model and its reactions were silenced. The flux of ATPasel reaction was then fixed to its maximum value as determined by FBA. Finally, we performed FVA for the knockout model to determine the new maximal and minimal values of each reaction flux [Hadi and Marashi Citation2014]. Similar to the work of Hadi and Marashi [Citation2014], we classified reactions into four groups by comparing the range of their flux in normal versus knockout states (see ). Furthermore, the DAVID functional annotation tool was used to find the positively influenced pathways in each knockout model [Huang et al. Citation2009].
Declaration of interest
Financial support of the University of Tehran for this research under Grant Number 28791/1/2 (S-AM). The authors report no confliction of interest and none of the authors of the paper are employed by the Government of Iran.
IAAN_1263367_Supplementary_Files.zip
Download Zip (262.1 KB)Acknowledgments
The authors would like to thank Dr. K. Gilany (Avicenna Research Institute, ACECR, Iran) for his assistance in data collection and helpful discussions. The updated Recon2 model was kindly provided by Dr. Nathan Lewis (UCSD). S-AM acknowledges the financial support of the University of Tehran for this research under Grant Number 28791/1/2.
Additional information
Notes on contributors
Arvand Asghari
Conceived and designed the experiments: S-AM, AA; Reconstructed the model, performed the computational experiments, and drafted the original report: AA; Involved in the interpretation of the findings and reviewing and revising the final manuscript: S-AM, AA, NAP.
Sayed-Amir Marashi
Conceived and designed the experiments: S-AM, AA; Reconstructed the model, performed the computational experiments, and drafted the original report: AA; Involved in the interpretation of the findings and reviewing and revising the final manuscript: S-AM, AA, NAP.
Naser Ansari-Pour
Conceived and designed the experiments: S-AM, AA; Reconstructed the model, performed the computational experiments, and drafted the original report: AA; Involved in the interpretation of the findings and reviewing and revising the final manuscript: S-AM, AA, NAP.
References
- Agarwal, A. and Said, T.M. (2004) Carnitines and male infertility. Reprod Biomed Online 8: 376–384.
- Agarwal, A., Sharma, R., Samanta, L., Durairajanayagam, D. and Sabanegh, E. (2016) Proteomic signatures of infertile men with clinical varicocele and their validation studies reveal mitochondrial dysfunction leading to infertility. Asian J Androl 18: 282–291.
- Amaral, A. and Ramalho‐Santos, J. (2010) Assessment of mitochondrial potential: implications for the correct monitoring of human sperm function. Int J Androl 33: e180–e186.
- Amaral, A., Castillo, J., Ramalho-Santos, J. and Oliva, R. (2013a) The combined human sperm proteome: cellular pathways and implications for basic and clinical science. Human Reprod Update 20: 40–62.
- Amaral, A., Lourenço, B., Marques, M. and Ramalho-Santos, J. (2013b) Mitochondria functionality and sperm quality. Reproduction 146: R163–R174.
- Amaral, A., Castillo, J., Estanyol, J.M., Ballescà, J.L., Ramalho-Santos, J. and Oliva, R. (2013c) Human sperm tail proteome suggests new endogenous metabolic pathways. Mol Cell Proteomics 12: 330–342.
- Amaral, A., Paiva, C., Attardo Parrinello, C., Estanyol, J.M., Ballescà, J.L., Ramalho-Santos, J., et al. (2014) Identification of proteins involved in human sperm motility using high-throughput differential proteomics. J Proteome Res 13: 5670–5684.
- Ashrafzadeh, A., Karsani, S.A. and Nathan, S. (2013) Mammalian sperm fertility related proteins. Int J Med Sci 10: 1649.
- Balercia, G., Buldreghini, E., Vignini, A., Tiano, L., Paggi, F., Amoroso, S., et al. (2009) Coenzyme Q 10 treatment in infertile men with idiopathic asthenozoospermia: a placebo-controlled, double-blind randomized trial. Fertil Steril 91: 1785–1792.
- Bansal, S.K., Gupta, N., Sankhwar, S.N. and Rajender, S. (2015) Differential Genes Expression between Fertile and Infertile Spermatozoa Revealed by Transcriptome Analysis. PloS One 10: e0127007.
- Bartellini, M., Canale, D., Izzo, P., Giorgi, P., Meschini, P. and Mechini-Fabris, G. (1986) L-carnitine and acetylcarnitine in human sperm with normal and reduced motility. Acta Europ Fertil 18: 29–31.
- Bordbar, A., Monk, J.M., King, Z.A. and Palsson, B.O. (2014) Constraint-based models predict metabolic and associated cellular functions. Nature Rev Genet 15: 107–120.
- Brown, G.C. (1995) Nitric oxide regulates mitochondrial respiration and cell functions by inhibiting cytochrome oxidase. FEBS Lett 369: 136–139.
- Cappello, A.R., Guido, C., Santoro, A., Santoro, M., Capobianco, L., Montanaro, D., et al. (2012) The mitochondrial citrate carrier (CIC) is present and regulates insulin secretion by human male gamete. Endocrinology 153: 1743–1754.
- Chan, C.-C., Shui, H.-A., Wu, C.-H., Wang, C.-Y., Sun, G.-H., Chen, H.-M., et al. (2009) Motility and protein phosphorylation in healthy and asthenozoospermic sperm. J Proteome Res 8: 5382–5386.
- Chi, A. and Kemp, R.G. (2000) The primordial high energy compound: ATP or inorganic pyrophosphate? J Biol Chem 275: 35677–35679.
- Costa, M., Canale, D., Filicori, M., D’lddio, S. and Lenzi, A. (1994) L‐carnitine in idiopathic asthenozoospermia: a multicenter study. Andrologia 26: 155–159.
- Dada, R., Mahfouz, R. Z., Kumar, R., Venkatesh, S., Shamsi, M. B., Agarwal, A., et al. (2011) A comprehensive work up for an asthenozoospermic man with repeated intracytoplasmic sperm injection (ICSI) failure. Andrologia 43: 368–372.
- De Rosa, M., Zarrilli, S., Paesano, L., Carbone, U., Boggia, B., Petretta, M., et al. (2003) Traffic pollutants affect fertility in men. Human Reprod 18: 1055–1061.
- Dirami, T., Rode, B., Jollivet, M., Da Silva, N., Escalier, D., Gaitch, N., et al. (2013) Missense mutations in SLC26A8, encoding a sperm-specific activator of CFTR, are associated with human asthenozoospermia. Am J Human Genet 92: 760–766.
- Dreanno, C., Cosson, J., Suquet, M., Seguin, F., Dorange, G. and Billard, R. (1999) Nucleotide content, oxydative phosphorylation, morphology, and fertilizing capacity. Mol Reprod Dev 53: 230–243.
- Du, Y., Li, M., Chen, J., Duan, Y., Wang, X., Qiu, Y., et al. (2015) Promoter targeted bisulfite sequencing reveals DNA methylation profiles associated with low sperm motility in asthenozoospermia. Human Reprod 31: 24–33.
- Duarte, N.C., Becker, S.A., Jamshidi, N., Thiele, I., Mo, M.L., Vo, T.D., et al. (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci 104: 1777–1782.
- Eslamian, G., Amirjannati, N., Rashidkhani, B., Sadeghi, M.-R., Baghestani, A.-R. and Hekmatdoost, A. (2015) Dietary fatty acid intakes and asthenozoospermia: a case-control study. Fertil Steril 103: 190–198.
- Ford, W. (2006) Glycolysis and sperm motility: does a spoonful of sugar help the flagellum go round? Human Reprod Update 12: 269–274.
- Ford, W. and Harrison, A. (1981) The role of oxidative phosphorylation in the generation of ATP in human spermatozoa. J Reprod Fertil 63: 271–278.
- Forgac, M. (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nature Reviews Mol Cell Biol 8: 917–929.
- Fouladiha, H., Marashi, S.A. and Shokrgozar, M. (2015) Reconstruction and validation of a constraint‐based metabolic network model for bone marrow‐derived mesenchymal stem cells. Cell Prolif 48: 475–485.
- Frezza, C., Zheng, L., Folger, O., Rajagopalan, K.N., MacKenzie, E.D., Jerby, L., et al. (2011) Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature 477: 225–228.
- Fukushima, T., Kaneoka, H., Yasuno, T., Sasaguri, Y., Tokuyasu, T., Tokoro, K., et al. (2013) Three novel mutations in the carnitine–acylcarnitine translocase (CACT) gene in patients with CACT deficiency and in healthy individuals. J Human Genet 58: 788–793.
- Golan, R., Weissenberg, R. and Lewin, L. (1984) Carnitine and acetylcarnitine in motile and immotile human spermatozoa. Int J Androl 7: 484–494.
- Goodrich, R.J., Anton, E. and Krawetz, S.A. (2013) Isolating mRNA and small noncoding RNAs from human sperm. Methods Mol Biol 927: 385–396.
- Gurunath, S., Pandian, Z., Anderson, R.A. and Bhattacharya, S. (2011) Defining infertility—a systematic review of prevalence studies. Human Reprod Update 17: 575–588.
- Hadi, M. and Marashi, S.-A. (2014) Reconstruction of a generic metabolic network model of cancer cells. Mol BioSystems 10: 3014–3021.
- Hinton, B., Snoswell, A. and Setchell, B. (1979) The concentration of carnitine in the luminal fluid of the testis and epididymis of the rat and some other mammals. J Reprod Fertil 56: 105–111.
- Huang, D.W., Sherman, B.T. and Lempicki, R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4: 44–57.
- Hwang, K., Walters, R.C. and Lipshultz, L.I. (2011) Contemporary concepts in the evaluation and management of male infertility. Nature Rev Urol 8: 86–94.
- Jerby, L., Shlomi, T. and Ruppin, E. (2010) Computational reconstruction of tissue‐specific metabolic models: application to human liver metabolism. Mol Syst Biol 7: 401.
- Jodar, M., Sendler, E. and Krawetz, S.A. (2016) The protein and transcript profiles of human semen. Cell Tissue Res 363: 85–96.
- John, J.C., Jokhi, R.P. and Barratt, C.L. (2005) The impact of mitochondrial genetics on male infertility. Int J Androl 28: 65–73.
- Johnson, G., Sendler, E., Lalancette, C., Hauser, R., Diamond, M.P. and Krawetz, S. (2011) Cleavage of rRNA ensures translational cessation in sperm at fertilization. Mol Human Reprod 17: 721–726.
- Kadenbach, B., Huttemann, M., Arnold, S., Lee, I. and Bender, E. (2000) Mitochondrial energy metabolism is regulated via nuclear-coded subunits of cytochrome c oxidase. Free Radic Biol Med 29: 211–221.
- Kamijo, T., Wanders, R., Saudubray, J., Aoyama, T., Komiyama, A. and Hashimoto, T. (1994) Mitochondrial trifunctional protein deficiency. Catalytic heterogeneity of the mutant enzyme in two patients. J Clin Invest 93: 1740.
- Larhlimi, A., David, L., Selbig, J. and Bockmayr, A. (2012) F2C2: a fast tool for the computation of flux coupling in genome-scale metabolic networks. BMC Bioinform 13: 57.
- Lewis, N.E., Schramm, G., Bordbar, A., Schellenberger, J., Andersen, M.P., Cheng, J.K., et al. (2010) Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nature Biotechnol 28: 1279–1285.
- Li, Y., Park, J.-S., Deng, J.-H. and Bai, Y. (2006) Cytochrome c oxidase subunit IV is essential for assembly and respiratory function of the enzyme complex. J Bioenerg Biomembranes 38: 283–291.
- Liu, F.-J., Liu, X., Han, J.-L., Wang, Y.-W., Jin, S.-H., Liu, X.-X., et al. (2015) Aged men share the sperm protein PATE1 defect with young asthenozoospermia patients. Human Reprod 30: 861–869.
- Luconi, M., Forti, G. and Baldi, E. (2006) Pathophysiology of sperm motility. Front Biosci 11: 1433–1447.
- Mahadevan, R. and Schilling, C. (2003) The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng 5: 264–276.
- Marchetti, C., Obert, G., Deffosez, A., Formstecher, P. and Marchetti, P. (2002) Study of mitochondrial membrane potential, reactive oxygen species, DNA fragmentation and cell viability by flow cytometry in human sperm. Human Reprod 17: 1257–1265.
- Mardinoglu, A. and Nielsen, J. (2012) Systems medicine and metabolic modelling. J Intern Med 271: 142–154.
- Mardinoglu, A., Agren, R., Kampf, C., Asplund, A., Uhlen, M. and Nielsen, J. (2014) Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nature Commun 5: 3083.
- Martínez-Heredia, J., de Mateo, S., Vidal-Taboada, J.M., Ballescà, J.L. and Oliva, R. (2008) Identification of proteomic differences in asthenozoospermic sperm samples. Human Reprod 23: 783–791.
- Mayr, J.A., Merkel, O., Kohlwein, S.D., Gebhardt, B.R., Böhles, H., Fötschl, U., et al. (2007) Mitochondrial Phosphate–Carrier Deficiency: A Novel Disorder of Oxidative Phosphorylation. Am J Human Genet 80: 478–484.
- Montjean, D., De La Grange, P., Gentien, D., Rapinat, A., Belloc, S., Cohen-Bacrie, P., et al. (2012) Sperm transcriptome profiling in oligozoospermia. J Assisted Reprod Genet 29: 3–10.
- Nayernia, K., Adham, I.M., Burkhardt-Göttges, E., Neesen, J., Rieche, M., Wolf, S., et al. (2002) Asthenozoospermia in mice with targeted deletion of the sperm mitochondrion-associated cysteine-rich protein (Smcp) gene. Mol Cell Biol 22: 3046–3052.
- Nishi, T. and Forgac, M. (2002) The vacuolar (H+)-ATPases—nature’s most versatile proton pumps. Nature Reviews Mol Cell Biol 3: 94–103.
- O’Brien, E.J., Monk, J.M. and Palsson, B.O. (2015) Using genome-scale models to predict biological capabilities. Cell 161: 971–987.
- Orth, J.D., Thiele, I. and Palsson, B.Ø. (2010) What is flux balance analysis? Nature Biotechnol 28: 245–248.
- Ota, K., Jaiswal, M.K., Ramu, S., Jeyendran, R., Kwak-Kim, J., Gilman-Sachs, A., et al. (2013) Expression of a2 vacuolar ATPase in spermatozoa is associated with semen quality and chemokine-cytokine profiles in infertile men. PloS One 8: e70470.
- Palmieri, F. (2013) The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol Aspects Med 34: 465–484.
- Palmieri, F. and Pierri, C.L. (2010) Mitochondrial metabolite transport. Essays Biochem 47: 37–52.
- Pascual, M., Cebrian-Perez, J., Lopez-Perez, M. and Muino-Blanco, T. (1996) Short-term inhibition of the energy metabolism affects motility but not surface properties of sperm cells. Biosci Rep 16: 35–40.
- Pereira, L., Gonçalves, J. and Bandelt, H. J. (2008) Mutation C11994T in the mitochondrial ND4 gene is not a cause of low sperm motility in Portugal. Fertil Steril 89: 738–741.
- Piomboni, P., Focarelli, R., Stendardi, A., Ferramosca, A. and Zara, V. (2012) The role of mitochondria in energy production for human sperm motility. Int J Androl 35: 109–124.
- Rezola, A., Pey, J., Rubio, Á. and Planes, F.J. (2014) In-silico prediction of key metabolic differences between two non-small cell lung cancer subtypes. PloS One 9: e103998.
- Roy, A., Lin, Y.-N., Agno, J.E., DeMayo, F.J. and Matzuk, M.M. (2007) Absence of tektin 4 causes asthenozoospermia and subfertility in male mice. FASEB J 21: 1013–1025.
- Ruiz-Pesini, E., Diez, C., Lapeña, A.C., Pérez-Martos, A., Montoya, J., Alvarez, E., et al. (1998) Correlation of sperm motility with mitochondrial enzymatic activities. Clin Chem 44: 1616–1620.
- Ruiz-Pesini, E., Lapena, A.-C., Díez-Sánchez, C., Pérez-Martos, A., Montoya, J., Alvarez, E., et al. (2000) Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am J Human Genet 67: 682–696.
- Ruiz‐Pesini, E., Díez‐Sánchez, C., López‐Pérez, M.J. and Enriquez, J.A. (2007) The role of the mitochondrion in sperm function: is there a place for oxidative phosphorylation or is this a purely glycolytic process? Curr Topics Dev Biol 77: 3–19.
- Ryu, J.Y., Kim, H.U. and Lee, S.Y. (2015) Reconstruction of genome-scale human metabolic models using omics data. Integr Biol 7: 859–868.
- Safarinejad, M.R., Safarinejad, S., Shafiei, N. and Safarinejad, S. (2012) Effects of the reduced form of coenzyme Q 10 (ubiquinol) on semen parameters in men with idiopathic infertility: a double-blind, placebo controlled, randomized study. J Urol 188: 526–531.
- Salazar, D.A., Rodríguez-López, A., Herreño, A., Barbosa, H., Herrera, J., Ardila, A., et al. (2016) Systems biology study of mucopolysaccharidosis using a human metabolic reconstruction network. Mol Genet Metabol 117: 129–139.
- Schellenberger, J., Que, R., Fleming, R.M., Thiele, I., Orth, J.D., Feist, A.M., et al. (2011) Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2. 0. Nature Protocols 6: 1290–1307.
- Sharpe, R.M. (2010) Environmental/lifestyle effects on spermatogenesis. Phil Transac R Soc Lond B Biol Sci 365: 1697–1712.
- Shen, S., Wang, J., Liang, J. and He, D. (2013) Comparative proteomic study between human normal motility sperm and idiopathic asthenozoospermia. World J Urol 31: 1395–1401.
- Shlomi, T., Cabili, M.N. and Ruppin, E. (2009) Predicting metabolic biomarkers of human inborn errors of metabolism. Mol Syst Biol 5: 263.
- Ślęzak, R., Szczepaniak, M., Pasińska, M. and Czemarmazowicz, H. (2007) The analysis of CFTR mutations in men with azoospermia, oligozoospermia and asthenozoospermia. Ginekol Pol 78: 605–610
- Sohrabi-Jahromi, S., Marashi, S.A. and Kalantari, S. (2016) A kidney-specific genome-scale metabolic network model for analyzing focal segmental glomerulosclerosis. Mamm Genome 27: 158–167.
- Song, P., Zou, S., Chen, T., Chen, J., Wang, Y., Yang, J., et al. (2015) Endothelial nitric oxide synthase (eNOS) T-786C, 4a4b, and G894T polymorphisms and male infertility: study for idiopathic asthenozoospermia and meta-analysis. Biol Reprod 92: 38.
- Sousa, A.P., Amaral, A., Baptista, M., Tavares, R., Campo, P.C., Peregrín, P.C., et al. (2011) Not all sperm are equal: functional mitochondria characterize a subpopulation of human sperm with better fertilization potential. PloS One 6: e18112.
- Storey, B.T. (2008) Mammalian sperm metabolism: oxygen and sugar, friend and foe. Int J Dev Biol 52: 427.
- Sun-Wada, G.-H., Imai-Senga, Y., Yamamoto, A., Murata, Y., Hirata, T., Wada, Y., et al. (2002) A proton pump ATPase with testis-specific E1-subunit isoform required for acrosome acidification. J Biol Chem 277: 18098–18105.
- Sun, J., Aluvila, S., Kotaria, R., Mayor, J.A., Walters, D.E. and Kaplan, R.S. (2010) Mitochondrial and plasma membrane citrate transporters: discovery of selective inhibitors and application to structure/function analysis. Mol Cell Pharmacol 2: 101.
- Swainston, N., Smallbone, K., Hefzi, H., Dobson, P.D., Brewer, J., Hanscho, M., et al. (2016) Recon 2.2: from reconstruction to model of human metabolism. Metabolomics 12: 109.
- Tanphaichitr, N. (1976) In vitro stimulation of human sperm motility by acetylcarnitine. International J Fertil 22: 85–91.
- Thiele, I., Swainston, N., Fleming, R.M., Hoppe, A., Sahoo, S., Aurich, M.K., et al. (2013) A community-driven global reconstruction of human metabolism. Nature Biotechnol 31: 419–425.
- Tomar, A.K., Saraswat, M., Chhikara, N., Kumar, S., Yadav, V.K., Sooch, B.S., et al. (2010) Differential proteomics of sperm: insights, challenges and future prospects. Biomarkers Med 4: 905–910.
- Tomar, R., Mishra, A.K., Mohanty, N.K. and Jain, A.K. (2012) Altered expression of succinic dehydrogenase in asthenozoospermia infertile male. Am J Reprod Immun 68: 486–490.
- Vernejoul, F., Ghenassia, L., Souque, A., Lulka, H., Drocourt, D., Cordelier, P., et al. (2006) Gene therapy based on gemcitabine chemosensitization suppresses pancreatic tumor growth. Mol Ther 14: 758–767.
- Visser, L., Westerveld, G.H., Xie, F., van Daalen, S.K., van der Veen, F., Lombardi, M.P., et al. (2011) A comprehensive gene mutation screen in men with asthenozoospermia. Fertil Steril 95: 1020–1024. e9.
- Wang, G., Guo, Y., Zhou, T., Shi, X., Yu, J., Yang, Y., et al. (2013) In-depth proteomic analysis of the human sperm reveals complex protein compositions. J Proteomics 79: 114–122.
- Wang, Y., Eddy, J.A. and Price, N.D. (2012) Reconstruction of genome-scale metabolic models for 126 human tissues using mCADRE. BMC Syst Biol 13: 153.
- Yi, Y.-J., Sutovsky, M., Kennedy, C. and Sutovsky, P. (2012) Identification of the inorganic pyrophosphate metabolizing, ATP substituting pathway in mammalian spermatozoa. PloS One 7: e34524.
- Yu, Q., Zhou, Q., Wei, Q., Li, J., Feng, C. and Mao, X. (2014) SEMG1 may be the candidate gene for idiopathic asthenozoospermia. Andrologia 46: 158–166.
- Zhang, C. and Hua, Q. (2015) Applications of genome-scale metabolic models in biotechnology and systems medicine. Frontiers Physiol 7: 413.
- Zhao, C., Huo, R., Wang, F.-Q., Lin, M., Zhou, Z.-M. and Sha, J.-H. (2007) Identification of several proteins involved in regulation of sperm motility by proteomic analysis. Fertil Steril 87: 436–438.
- Zuccarello, D., Ferlin, A., Garolla, A., Pati, M.A., Moretti, A., Cazzadore, C., et al. (2008) A possible association of a human tektin-t gene mutation (A229V) with isolated non-syndromic asthenozoospermia: case report. Human Reprod 23: 996–1001.
