Abstract
Male infertility is a reproductive disorder, accounting for 40–50% of infertility. Currently, in about 70% of infertile men, the cause remains unknown. With the introduction of novel omics and advancement in high-throughput technology, potential biomarkers are emerging. The main purpose of our work was to overview different aspects of omics approaches in association with idiopathic male infertility and highlight potential genes, transcripts, non-coding RNA, proteins, and metabolites worth further exploring. Using the Gene Ontology (GO) analysis, we aimed to compare enriched GO terms from each omics approach and determine their overlapping. A PubMed database screening for the literature published between February 2014 and June 2022 was performed using the keywords: male infertility in association with different omics approaches: genomics, epigenomics, transcriptomics, ncRNAomics, proteomics, and metabolomics. A GO enrichment analysis was performed using the Enrichr tool. We retrieved 281 global studies: 171 genomics (DNA level), 21 epigenomics (19 of methylation and two histone residue modifications), 15 transcriptomics, 31 non-coding RNA, 29 proteomics, two protein posttranslational modification, and 19 metabolomics studies. Gene ontology comparison showed that different omics approaches lead to the identification of different molecular factors and that the corresponding GO terms, obtained from different omics approaches, do not overlap to a larger extent. With the integration of novel omics levels into the research of idiopathic causes of male infertility, using multi-omic systems biology approaches, we will be closer to finding the potential biomarkers and consequently becoming aware of the entire spectrum of male infertility, their cause, prognosis, and potential treatment.
Introduction
Infertility is a disease of the reproductive system defined by the inability to conceive after at least one year of regular, unprotected sexual intercourse (Zegers-Hochschild et al. Citation2009). Infertility affects 15% of couples in their reproductive age and a male factor is estimated to contribute to 50% of cases (Dohle et al. Citation2005; Agarwal et al. Citation2015). There is a wide range of causes associated with male infertility. It can be due to congenital factors, which include genetic disorders and chromosome abnormalities, and acquired factors like endocrine disorders, infections, tumors, injuries, toxins, and even circadian rhythm disruptions like seasonal changes and sleep/wake cycles can influence the quality of semen (Leaver Citation2016; Peterlin et al. Citation2019).
Male infertility can also be due to genetic disorders like Klinefelter syndrome (Fainberg et al. Citation2019), microdeletions of the Y chromosome (Liu et al. Citation2016), or it can be just an additional phenotype in syndromes, like deafness‐infertility syndrome, Kartagener syndrome, and primary ciliary dyskinesia (Mikec et al. Citation2022). Other genetic causes include chromosome abnormalities, like the formation of ring chromosomes or translocations (Sobotka et al. Citation2015; Barišić et al. Citation2021; Berkay et al. Citation2023). However, in 60–75%, the cause of male infertility remains unknown, also termed idiopathic male infertility (Punab et al. Citation2017). In recent years, a lot of effort has been made toward the diagnosis of idiopathic male infertility. Despite years of research, little is known about the identification of recurrent genetic factors, biomarkers, and potential clinical applications (Krausz et al. Citation2015).
Idiopathic male infertility is caused by the interaction of genetic and environmental factors (Jungwirth et al. Citation2012). Therefore, a high proportion of cases with idiopathic male infertility have been left to search for various causes originating in novel omics, including genomics, epigenomics, transcriptomics, proteomics, metabolomics, and miRNAomics/ncRNAomics.
In recent years, the use of next-generation squencing (NGS) methods, including whole exome and genome sequencing (WES and WGS) has increased exponentially in research and diagnostic settings. Although WGS appears to detect copy number variations as well as non-coding and intergenic regions, due to lesser cost whole exome sequencing is the more favorable approach in diagnostic and clinical settings (Pirih and Kunej Citation2017). In addition to genomics, new technologies, and lower costs have also led to an expansion of research in other omics disciplines (Dai and Shen Citation2022). Each of the listed omics approaches individually or in combination with other omics approaches can help identify and characterize new molecules involved in male infertility (Hasin et al. Citation2017).
Some attempts have been made to provide an overview of the field of omics research on male infertility (Llavanera et al. Citation2022; Omolaoye et al. Citation2022; Wagner et al. Citation2023) but because of the extensive research on male infertility in recent years and the increasing amount of data, new, up-to-date reviews of this field are needed. Nonetheless, omics research also presents some challenges. The common challenge of all approaches is that they generate large datasets, so noise can overwhelm the signal, which can reduce the sensitivity (Ning and Lo Citation2010). A comprehensive approach is needed for evaluation to determine whether the results of the different omics approaches are comparable or rather to find commonly enriched mechanisms.
Therefore, due to the high complexity of male infertility and the lack of reviews on multi-omics approaches in the field of male infertility, we provide an omics-based systematic review of male infertility, that includes recent data from 2014 to 2022. The aim was to analyze different omics studies to provide an overview of the field of male infertility and to gain insight into emerging evidence. We performed a Gene Ontology (GO) enrichment analysis for each sample type separately and compared the omics approaches to answer if different omics approaches access the same molecular factors.
Review and data synthesis
Literature search
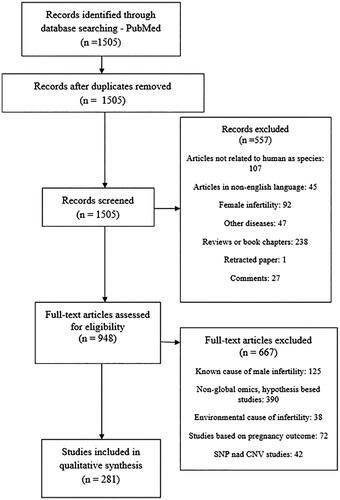
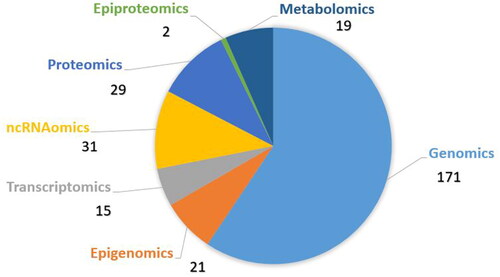
The literature screening resulted in 1505 studies. Using the strategy outlined in section ‘Methods’, 1224 studies were excluded. In the analysis, we included 281 studies that investigated different types of omics and idiopathic male infertility (). Some studies covered different omics approaches, so we included these studies in more than one omics section. One hundred and seventy-one genomics studies of rare deleterious sequence variants, 21 studies on epigenomics (19 on methylation and two on histone modifications), 15 studies on transcriptomics, 31 studies on ncRNAomics, 29 studies on proteomics, two on epi-proteomics – protein posttranslational modifications, and 19 studies of metabolomics were obtained (). Male infertility in the studies was characterized by the terms oligozoospermia, asthenozoospermia, teratozoospermia, a combination of them – oligoasthenoteratozoospermia, nonobstructive azoospermia, obstructive azoospermia, infertile normozoospermic, idiopathic infertility or just undefined male infertility. We extracted the highlighted molecular factors from the global studies. All data extracted from the literature and the list of 281 references with PMID numbers are presented in Supplementary Tables S1–S24.
Table 1. Rare sequence variants in genes associated with male infertility.
Genomics – Rare sequence variants
We have collected published studies, which used the WES or WGS technologies for the identification of variants associated with male infertility in humans. The most studied causes of male infertility were quantitative sperm abnormalities (non-obstructive/obstructive azoospermia and oligozoospermia), followed by motility (asthenozoospermia) and morphological abnormalities (teratozoospermia). All studies were divided based on different types of sperm abnormalities into three groups: quantitative, motility, and morphological abnormalities. The sample type used in genomics studies was determined to be blood.
We obtained 77 studies associated with quantitative sperm abnormalities (Supplementary Table S1), which included 144 genes (). In most studies, the main methodology, including combinations with other methods, was WES (n = 76). Some studies included different methods. WGS was performed in only one study. There were 21 studies with sporadic cases and 40 studies with familiar cases. Four studies were case studies and 12 were a combination of familiar and sporadic cases. More than half of the studies included men with non-obstructive azoospermia (n = 52). Of the 144 genes, 37 of them were reported in more than one study (). Two of them: ADGRG2 and CFTR were involved with obstructive azoospermia.
We further obtained 61 studies related to male infertility with motility sperm defects (Supplementary Table S2) and retrieved 44 genes (). Here as well WES, included in combinations, was the most common methodology (n = 60), followed by WGS (n = 1). There were 33 studies with sporadic cases and 20 studies with familiar cases. We collected one case study and seven studies, which were a combination of familiar and sporadic studies. Patients in 45 studies had, besides asthenozoospermia, multiple morphological abnormalities of the sperm flagella (MMAF). Of the 44 obtained genes, 14 of them were reported with asthenozoospermia in more than one study ().
Among idiopathic infertile men with morphological sperm abnormality, 33 studies were obtained (Supplementary Table S3). In this group of infertile men, 34 genes were retrieved (). All studies were performed using WES. We obtained 15 studies with sporadic cases and 13 familiar studies. Five studies had both sporadic and familiar cases. The phenotypes were listed as teratozoospermia, globozoospermia, or acephalic spermatozoa syndrome in combination with MMAF or other morphological defects like absent acrosome. Nine studies had cases with MMAF, and in four of them, the phenotype was globozoospermia. Out of 34 genes, eight were reported with sperm morphological abnormalities in more than one study ().
There were 42 genes involved with MMAF, which leads to both asthenozoospermia and teratozoospermia. In Supplementary Tables S2 and S3, we classified them according to the phenotype mentioned in the corresponding study. These involved genes were: CCDC34, AK7, DNAH1, SPAG17, CFAP43, CFAP44, CFAP69, CFAP251, QRICH2, ARMC2, TTC21A, SPEF2, CFAP65, CFAP70, DNAH17, DNAH6, TTC29, DZIP1, CFAP91, WDR19, DNAH8, CFAP58, CFAP47, IFT74, DNAH2, DNAH12, DRC1, MDC1, PACRG, SPPL2C, TPTE2, STK33, DNAH10, FSIP2, SPAG6, CEP135, PIWIL4, CC2D1B, CCNB3, KIAA1210, CHPT1 and SEPTIN12.
Table 2. Differentially methylated genes associated with male infertility.
Table 3. Differentially expressed genes associated with male infertility.
Seven genes (DNAH6, USP26, CFAP44, DZIP1, TTC21A, SPAG17, and SLC26A8) were observed in both the quantitative and motility sperm abnormalities, two (BRDT and FBXO43) in both quantitative and morphological sperm abnormalities and seven in both (DNAH2, CFAP58, FSIP2, SPEF2, DNAH10, DNAH17, and DNAH12) motility and morphological sperm abnormalities. Variants in DNAH1 were however observed in all three sperm abnormalities.
Overall, 171 studies reporting 204 genes related to genetic causes of unexplained male infertility were obtained (Supplementary Tables S1–S3).
A GO analysis for genes involved with quantitative, motility, and morphological sperm abnormalities was performed. Categorizing the genes, based on the type of sperm abnormality, we found that the most enriched biological processes in the quantitative sperm abnormality group were homologous chromosome pairing at meiosis, female gamete generation, and synaptonemal complex assembly. The most enriched terms for the GO category molecular function were MutSalpha complex binding and DNA binding, and for cellular component it was chromosome.
In the motility and morphological sperm abnormalities, the most enriched biological processes were cilium movement and axonemal dynein complex assembly, and cilium organization. The most enriched molecular functions were beta-tubulin binding and CDP-alcohol phosphatidyltransferase activity and for the cellular component; the 9 + 2 motile cilium and sperm flagellum.
The GO analysis of all obtained genes is presented in the Supplementary Figures S1–S4. The 10 most enriched terms for the input gene set are shown based on the –log10(p value), with the actual p value next to each term. The term at the top has the most significant overlap with the input query gene set. An asterisk (*) next to a p value indicates a significantly adjusted p value (<.05) for that term. Adjusted p values were calculated using the Benjamini–Hochberg method as indicated by Enrichr.
Epigenomics
From the epigenomics field, we obtained studies, which included the association of aberrant methylation of several genes and alterations of posttranslational modifications in histones among infertile men. We retrieved more studies with an aberrant methylation of genes in association with male infertility, compared to the alterations of posttranslational modifications in histones.
Nineteen studies reporting the association between aberrant methylation and male infertility were obtained (Supplementary Tables S4–S6). Altogether 829 men were recruited as infertile, and 533 as healthy controls. The most frequently observed sample used was sperm (n = 13), followed by testicular tissue (n = 4) and blood (n = 2). For the methods, the most frequently used were methylation arrays (n = 14) and bisulfite sequencing (n = 4, and an additional 12 studies for validation). One study was done using methylated DNA immunoprecipitation coupled with NGS. Genes or gene-related amplicons from the global studies are presented in .
Table 4. Differentially expressed ncRNAs associated with male infertility.
The GO analysis of genes found differentially methylated in infertile males, showed the most enriched biological processes to be positive regulation of morphogenesis of an epithelium (blood samples), regulation of oxidative stress-induced cell death (sperm samples), and 3-UTR-mediated mRNA stabilization and piRNA processing (testicular tissue samples). The GO analysis of obtained genes for each sample type, including molecular function and cellular component enrichment analysis is presented in Supplementary Figures S5–S7.
We additionally retrieved two studies of histone residue modifications. The studies were focused on histones H3 and H4. A total of 81 men; 47 infertile and 34 controls were recruited. Acetylation and methylation alterations were observed on H3 and H4 and S-sulfhydration changes on H3. Both studies were performed on sperm with LC-MS/MS methodology (Supplementary Table S7).
Transcriptomics
We retrieved 15 global transcriptome studies (Supplementary Tables S8–S10). Altogether, 474 infertile and 160 controls were recruited in the studies. The most used methodology in the studies were microarrays (n = 8) and RNA-sequencing (n = 7 including one single-cell RNA-sequencing). The validation of global data was done with the use of qPCR. The most used sample was testicular tissue (n = 7, of which two were done on Sertoli cells), followed by sperm (n = 6) and seminal plasma (n = 2). Genes and their transcripts from the global studies are presented in .
GO analysis of the obtained transcripts, differentially expressed in infertile men, showed that the most enriched biological processes were cytoplasmic translation (sperm samples), metanephros development, and positive regulation of development process (testicular tissue samples) and synaptonemal complex assembly (seminal plasma samples). The GO analysis of obtained genes/transcripts for each sample type, including the results of the molecular function and cellular component enrichment analysis is presented in Supplementary Figures S8–S10.
Non-coding RNA omics (ncRNAomics)
Thirty-one global ncRNAomics studies in association with male infertility were obtained (Supplementary Tables S11–S15). Eighteen studies were related to miRNA, five studies with lncRNA, four studies with piRNA, two studies with circRNA, one study with both miRNA and piRNA, and one with both lncRNA and miRNA.
Altogether 1355 infertile men and 671 controls were recruited in the studies. For two studies, the number of participants was not provided in the article (Tables S11–S15). The most common sample analyzed was testicular tissue (n = 12, of which one was on Sertoli cells), followed by seminal plasma (n = 10), sperm (n = 7), blood (n = 1), and both seminal plasma and testicular tissue (n = 1). The leading methodologies were RNA sequencing and microarrays. Most of the studies validated their results with qPCR. Fourteen studies were done using RNA sequencing, 11 with microarrays, four with qPCR arrays, and one each with TaqMan Low-Density Array and PCR panels of 742 miRNA.
The ncRNAs that were investigated and were found to be differentially expressed in infertile men compared to fertile are presented in .
Proteomics
We obtained 29 global proteomics studies (Supplementary Tables S16–S19). The studies recruited 572 men as cases of male infertility and 415 men as controls. The most commonly used methods in global studies were various combinations of mass spectrometry (MS) (n = 28). In addition, two-dimensional differential gel electrophoresis (2D-DIGE) and 4D quantitative proteomic analysis-trapped ion mobility spectrometry were used as proteomic methods (Supplementary Tables S16–S19). The most frequently used sample was sperm (n = 14), followed by seminal plasma (n = 10), testicular tissue (n = 4), and both seminal plasma and sperm (n = 1). The proteins from the global studies are listed in .
Table 5. Differentially expressed proteins associated with male infertility.
Table 6. Differentially abundant metabolites associated with male infertility.
GO analysis of the obtained proteins, differentially expressed in infertile men, showed that the most enriched biological processes were glycolytic/carbohydrate catabolic process (sperm samples), CRD-mediated mRNA stabilization, and negative regulation of nuclear-transcripted mRNA catabolic process, deadenylation-depended decay (testicular tissue samples) and retina homeostasis and negative regulation of peptidase activity (seminal plasma samples). The GO analysis of the obtained proteins for each sample type, including the results of the molecular function and cellular component enrichment analysis is presented in Supplementary Figures S11–S13.
Two studies of posttranslational modifications of proteins, belonging to the category of epi-proteomics, were also obtained (Supplementary Table S20). One study involved lysine glutarylation and the other lysine 2-hydroxyisobutyrylation. In both studies, the infertile men had asthenozoospermia. A total of 124 men with asthenozoospermia and 119 controls participated in the studies. Both studies were performed on sperm using immunoblotting and immunofluorescence assays.
Metabolomics
Nineteen global metabolomic studies associated with male infertility were obtained (Supplementary Tables S21–S24). A total of 1646 men with male infertility and 1188 healthy controls were recruited for the studies. Four different sample types were used in the studies: seminal plasma (n = 12), followed by blood (n = 3), urine (n = 3), and sperm (n = 1). The most common method was MS (n = 14), followed by 1H NMR spectroscopy (n = 4) and Raman spectroscopy (n = 1). The metabolites differentially abundant in infertile men compared with fertile men are listed in .
Comparison of omics approaches
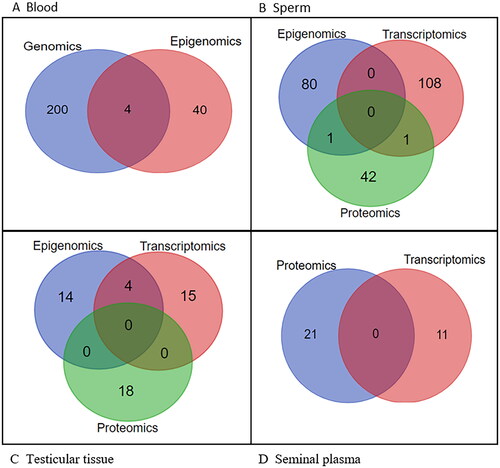
A Venn diagram showing the overlapping components of the various omics approaches by sample type revealed no significant overlap (). However, most overlap was between genomics and epigenomics in blood samples (DNAH17, ELMO1, PDHA2, PIWIL2), and epigenomics and transcriptomics (BOLL, DDX4, HORMAD1, MAEL) in testicular tissue samples. No overlap was observed in seminal plasma samples.
Discussion
The present study reviews recent non-hypothesis-based studies of male infertility in relation to different global omics approaches. We described the main approaches used in omics research and some typical studies in the field. The most frequently studied omics approach was genomics (rare sequence variants), followed by ncRNAomics, proteomics, epigenomics-methylation, metabolomics, and transcriptomics. In addition, two studies on histone residue modifications and protein posttranslational modifications were obtained (detailed in Supplementary Tables S1–S24).
Genomics
It is estimated that more than 1000 genes are testis-enriched, but only a few have been associated with male infertility (Djureinovic et al. Citation2014). The genes in our review, that were obtained in more than one study, may be of interest for future research and have diagnostic potential due to their reproducibility in infertile men. Despite the large number of genes in the quantitative sperm abnormalities group, some of them have been additionally associated with motility and morphological abnormalities in the literature: CCDC146, CEP131, CFAP44, DNAH1, DNAH6, DNAH7, ODF4, PGK2, SLC26A8, SPAG17, TTC21A and TTLL9 (Danshina et al. Citation2010; Hall et al. Citation2013; Konno et al. Citation2016; Coutton et al. Citation2018; Xu et al. Citation2018; Liu, He, et al. Citation2019; Tu et al. Citation2019; Gao et al. Citation2021; Wei et al. Citation2021; Martinez et al. Citation2022; Ito et al. Citation2023; Wang et al. Citation2023). The presence of rare deleterious variants in these genes in patients with quantitative sperm abnormalities suggests that the genes involved in male infertility remain understudied and that more research is needed to identify the mechanisms leading to male infertility.
Previous studies have found ‘de novo’ mutations in genes involved in male infertility and demonstrated their causality (Hodžić et al. Citation2021; Oud et al. Citation2022). We would therefore expect such an event to occur ‘de novo’ if we assume that infertile men cannot father children. Interestingly, about half of the studies that identified genes in our review were found in familial cases. In reviewing the data obtained, many patients were found to carry rare variants in homozygous or compound heterozygous form. Autosomal recessive inheritance of these variants was more common in consanguineous families (Sha et al. Citation2018; Shen et al. Citation2019; Jaillard et al. Citation2020; Li et al. Citation2022). In addition, transmission in these cases of male infertility could also occur using the help of artificial reproductive techniques (Silber and Repping Citation2002). Knowledge of genetic mutation could be used in artificial reproduction techniques using preimplantation genetic testing for monogenic disorders (PGT-M). This could be used to select embryos that do not carry the same mutation (Lee et al. Citation2017).
It is noteworthy that the study of genomics in the context of infertility has been the subject of extensive research. A recent review noted an increasing number of high-probability genes for male infertility genes and identified 104 high-probability genes associated with the aforementioned condition (Houston et al. Citation2021). Although more new genes related to male infertility are being discovered with the help of NGS technology, these genes remain primarily of diagnostic interest (Krzastek, Smith, et al. Citation2020).
Transcriptomics
Of all the omics approaches, transcriptomics was the least represented. The inclusion of studies based on the GEO database would likely result in a higher number. Transcriptomic assays provide a better understanding as they analyze the expression profile of different genes (Garrido and Hervás Citation2020). New technologies such as RNA-sequencing provide better resolution and higher coverage of the transcriptome compared to microarrays and can identify novel transcripts or alternative splice variants (Marioni et al. Citation2008; Kukurba and Montgomery Citation2015). However, notwithstanding the great potential of studying mRNA, the majority of transcripts are represented by non-coding RNAs (Seal et al. Citation2020).
Epigenomics and ncRNAomics
DNA methylation as a marker of infertility is an attractive target for studying epigenomics changes because it is the most robust epigenetic mark compared to histone modifications and RNA expression studies, which require a more careful storage approach (Mikeska and Craig Citation2014). Environmental components, such as plastic have also been linked to methylation changes (Manikkam et al. Citation2013). For this reason, research on environmental factors affecting sperm quality and male infertility has increased (Krzastek, Farhi, et al. Citation2020). Although epigenetics includes changes in DNA methylation, histone modifications, nucleosome positioning, and ncRNA, which we analyzed separately, we concluded that the most studied area, except ncRNA, is still DNA methylation. In addition, we found numerous studies on ncRNAs and male infertility, including most in the area of miRNAs. We obtained numerous ncRNAs, such as miRNA, lncRNA, piRNA, and circRNA down-regulated or up-regulated in men with infertility, of which some miRNAs were in the literature purposed as potential therapeutic targets (Rastgar Rezaei, et al. Citation2021), for example for potential contraceptives (Khazaie and Nasr Esfahani Citation2014).
Proteomics
Proteins are better indicators of the current state of the cell compared to genes because an expressed gene is not necessarily translated into a protein. For this reason, there are many studies targeting proteins as biomarkers. The disadvantage of proteomics is that proteins vary between different cells and can be affected by environmental factors, making them difficult to use in diagnostics. In addition, proteins in semen are a mixture of epididymis, prostate, seminal vesicles, and other glands (Krzastek, Smith, et al. Citation2020). Numerous obtained global proteomics studies indicate great interest and growth in this approach.
Metabolomics
Genomics and proteomics can detect altered components of the metabolic pathway, but non-invasive metabolomics has many advantages when applied in the clinical setting (Zhang et al. Citation2014). Because metabolites are the end product of genes, they are better at representing the cellular state than other omics approaches (Krzastek, Smith, et al. Citation2020). Unfortunately, there are also some drawbacks, such as poor reproducibility of some studies (Blaurock et al. Citation2022). Improving metabolomics methods could lead to the identification of novel biomarkers, as some metabolites are already known to correlate with sperm parameters (Blaurock et al. Citation2022). The literature review revealed that the most commonly studied samples in all omics approaches were sperm and testicular tissue, with the exception of the metabolomics where the leading sample was seminal plasma.
In addition to the aforementioned omics approaches, microbiomics, and its subfields are also gaining interest. Comparing idiopathic infertile and fertile men, some differences in several key bacterial and metabolic pathways were identified, representing the potential for diagnosis and treatment in the future (Lundy et al. Citation2021). This also highlights the complexity of idiopathic infertility and points to a new direction for research on male infertility.
Gene Ontology
Comparison of GO analysis revealed differences between genes involved in quantitative sperm abnormalities and genes involved in morphological and motility sperm abnormalities. As expected, in the quantitative sperm abnormalities, the most enriched process was meiosis, and in the other two, cilium movement and spermatid development. The similarity between the processes in asthenozoospermia and teratozoospermia may be due to a large number of genes involved in MMAF, that can lead to both conditions.
In terms of molecular function, the most enriched term in quantitative sperm abnormalities was MutSα complex binding. MutSα is mainly involved in mismatch repair (Edelbrock et al. Citation2013). STRING analysis (Szklarczyk et al. Citation2019) revealed that MLH1, MCM8, and MCM9 are annotated to the MutSα pathway. In addition to their role in replication initiation, MCM8 and MCM9 are also involved in homologous repair in somatic cells and during gametogenesis, as men with a variant in MCM8 were infertile and showed impaired repair of chromosome breaks (Lutzmann et al. Citation2012; Tenenbaum-Rakover et al. Citation2015). In addition, MLH1 plays a role in mismatch repair but is also involved in meiosis (Hunter and Borts Citation1997). In asthenozoospermia, the term beta-tubulin binding is also consistent with its involvement in motility, as IFT74 variants have been associated with skeletal ciliopathy and motile cilia abnormalities (Bakey et al. Citation2023). CHPT1 has been associated with the enriched molecular function term CDP-alcohol phosphatidyltransferase activity in teratozoospermia. The role of CHPT1 was associated with sperm head development, as abnormalities led to globozoospermia (Li et al. Citation2021). This indicates that male infertility is complex and that genes associated with specific processes and functions have additional roles in other processes that remain to be discovered.
Differences were found when comparing the GO profile of genomics, epigenomics, transcriptomics, and proteomics data. Due to the inclusion of studies performing the analysis on different sample types, the GO analysis and omics comparison was performed for each sample type separately. Comparison regardless of sample type would introduce additional noise, which would hinder the accurate analysis and therefore the reliability of obtained results (Misra et al. Citation2018).
Differences in GO terms between different sample types were observed in all non-genomics approaches. Regardless of the sample type, protein or regulation-involved processes were observed. Except for genomics, GO analysis of epigenomics, transcriptomics, and proteomics has resulted in mainly non-reproduction-specific processes, functions, and components. This may be due to the inclusion of only genes, transcripts, and proteins highlighted in the global studies, as global omics studies may lead to the identification of >1000 components. Compared to the genomics level, participants in other omics levels were also not grouped by male infertility phenotype. Another explanation is that the field of genomics is still the most researched so the functional involvement in male infertility is better known.
From the blood samples in the epigenomics approach, the most enriched terms: positive regulation of morphogenesis of an epithelium and regulation of epithelial cell differentiation involved with kidney development initially did not seem to be associated with infertility; however, a literature search of genes involved with the terms resulted in the opposite. One of those genes was PAX8. Even though PAX8 is involved with the proper development and differentiation of thyroid follicular cells (Di Palma et al. Citation2013), animal studies have observed its involvement with infertility, as Pax8-deficient mice were infertile, due to the absence of efferent ducts and epididymides or reduced efferent duct lumen, leading to the absence of spermatozoa in epididymis (Wistuba et al. Citation2007).
Non-specific and non-reproduction-related processes and molecular functions have also been observed in transcriptomics, with the highest enrichment in cytoplasmic translation for sperm samples. For example, RPL30 (60S ribosomal protein L30) was shown to be differentially expressed in asthenozoospermic men. The ribosomal abnormalities could affect ribosomes in the mitochondria in sperm, which in turn could affect motility (Bansal et al. Citation2015). The same non-specificity was observed in proteomics data from sperm samples with the most enriched term being glycolytic process. One of these genes involved in glycolysis are GAPDHS and PGK2, which are specifically expressed in most meiotic germ cells and are located in the tail of spermatozoa (Liu, Li, et al. Citation2019). The non-reproduction-specific processes and functions, observed in our results suggest a broad spectrum of molecular pathways involved in the infertility phenotype.
Analysis of the Venn diagram revealed no significant overlap between the obtained molecular factors of the aforementioned omics approaches, which could be due to focusing only on the highlighted molecular factors. Despite the aim of our study to analyze only the highlighted molecular factors, the study still provides the basis for future larger bioinformatics studies.
Nevertheless, a small overlap was found in blood samples between genomics and epigenomics (DNAH17, ELMO1, PDHA2, PIWIL2), testicular tissue samples between epigenomics and transcriptomics (BOLL, DDX4, HORMAD1, MAEL), sperm samples between epigenomics and proteomics (ANXA2) and transcriptomics and proteomics (SEMG1). No overlap was observed in seminal plasma samples. This could be due to a small number of obtained molecular factors in seminal plasma studies. On this basis, our results show that different omics approaches lead to different results or reveal different mechanisms and that further research is needed.
As mentioned above, male infertility is very heterogeneous and manifests in different phenotypes, ranging from normozoospermic infertility to testicular insufficiency without spermatozoa. Because spermatogenesis itself is a complex process, many genes and their products are involved. New data are emerging rapidly, as genetic variants and animal studies in novel genes beyond our search limit have already been associated with male infertility, for example, DNALI1 with asthenoteratozoospermia (Sha et al. Citation2022; Wu et al. Citation2023; Yap et al. Citation2023), MEIG1 with sperm motility in idiopathic infertility (Zhang et al. Citation2009; Li et al. Citation2015; Gupta et al. Citation2022) and KCTD19 with non-obstructive azoospermia (Liu et al. Citation2023). Further research is needed to classify the role of some factors in male infertility.
Given the large amount of data obtained from global studies, an integration of all different omics approaches will be required in the future. Systems biology approaches that incorporate other omics levels, such as microbiomics, and capture and integrate global datasets of different types, will help discover underlying mechanisms and putative biomarkers. New development of computational tools will accelerate the process (Aderem Citation2005; Zupanic et al. Citation2020).
Limitations of the study
Although the literature synthesis of the present study has contributed to the development of the field, there are also some limitations. Since this is a systematic review, we limited our search to the publication date in the last few years, which might lead us to miss some molecular factors. Another limitation is that the GO analysis was not performed for all identified molecular factors from global studies, because the number of most reached >100 or even 1000. Due to the large heterogeneity of infertility phenotypes or studies including multiple infertility phenotypes in the same publication, the analysis of epigenomics, transcriptomics, and proteomics data was not grouped based on the phenotype. Because of the large heterogeneity of ncRNAomics results and the inability of the Enrichr tool to provide a GO analysis for ncRNAs, their elimination could lead us to miss certain biological processes, molecular functions, and cellular components. Moreover, because the recovered ncRNAs target many genes, the analysis would be very extensive and should be the subject of future research.
Future directions
Due to the small number of studies obtained for some omics approaches and the numerous phenotypes, GO analysis on larger datasets is recommended, as well as the categorizing for infertility phenotype, which would lower the level of noise. Similarly, miRNAs should be included in pathway enrichment analysis. Another suggestion is to adopt a system that categorizes abnormalities according to the level of evidence for involvement in male infertility.
Conclusions
Several promising molecular factors have been obtained from retrieved global omics studies. Nevertheless, the path to discovering the ideal biomarkers for idiopathic male infertility is far more complex and opens the doors for personalized medicine, as idiopathic male infertility is associated with heterogeneous complex phenotypes. Our results show that each omics approach is associated with a different molecular profile, which complicates the identification of reliable biomarkers. In our work, we summarized the current knowledge on the molecular aspect of male infertility and showed which omics approaches are still under-researched, and where further research is needed. Further omics studies of idiopathic male infertility and their integration are needed for better diagnosis, prognosis, and potential therapy of male infertility. Current studies should focus on investigating predictive biomarkers for idiopathic male infertility based on the integration of multiple omics and systems biology approaches as male infertility is a complex biological system that should not be oversimplified. With new research incorporating new literature and new genes, we will move closer to understanding the mechanisms of male infertility.
Methods
PubMed literature screening
A PubMed database was screened, using the keywords ‘male infertility’ AND ‘WES’ OR ‘WGS’ OR ‘exome sequencing’ OR ‘transcriptomics’ OR ‘metabolomics’ OR ‘epigenomics’ OR ‘methylation’ OR ‘histone modification’ OR ‘proteomics’ OR ‘transcriptomics’ OR ‘mRNA’ OR ‘miRNA’ OR ‘piRNA’ OR ‘lncRNA’. All articles were related to humans as species and were manually extracted for relevant information. For each study, we obtained PMID IDs, cohort and cohort size, methodology, and the result of a study; potential molecular factors of idiopathic male infertility were reported such as genes, transcripts, epigenetic markers, proteins, ncRNAs, and metabolites. According to a potential molecular factor, relevant information was gathered: sequence variant, up/down-regulation of genes, gene methylation status, gene/protein expression, metabolites, and ncRNA expression. For cohort reporting, we included sporadic or familial cases. The term case was used when no family member sequencing was available. In some studies, combinations of sporadic and familial studies were found. The collected data were complemented with additional information: marker symbols and names were unified using HUGO Gene Nomenclature Committee (HGNC) for genes (Seal et al. Citation2023). If the symbol of the gene or gene product in the study had not been the same as observed on the HGNC, we included the latest approved gene symbol. We included the new approved gene symbol and the previous or alias symbols in the bracket.
Inclusion and exclusion criteria of studies for the qualitative synthesis
The inclusion criteria for this review were all studies related to idiopathic male infertility with the restriction to the publication date within the last years from February 2014 to June 2022. We included studies with only male infertility as well as studies, whose aim was to compare groups of male infertility and healthy controls. We focused only on the studies, which were done with omics technologies. Many genomic studies have identified variants by combining different methods. We, therefore, included all observed variants from studies, to avoid missing data, but only if the primary method was WES or other omics technology.
The exclusion criteria of studies were non-English language and known causes of male infertility (chromosomal abnormalities, genetic diseases, congenital and acquired abnormalities). Studies exploring men’s infertility connected to pregnancy outcome and DNA integrity were also excluded, because of the many variables, which can influence the phenotype. Studies in which they used only targeted exome sequencing, Sanger sequencing, or other non-NGS sequencing methods and ones in which the experimental data did not confirm the causality of the gene or were contrary, were excluded from this review. We also excluded infertile men who had additional health issues, like primary ciliary dyskinesia or diagnoses, which are known to potentially affect infertility (varicocele). Studies in which the transcriptome analysis was done with data downloaded from the gene expression omnibus (GEO) database were excluded in this example review, as for studies that explored individual genes/transcripts/non-coding RNA and proteins and were hypothesis-based. For the genomic studies (rare sequence variants), we only extracted variants present in affected infertile men. The present review was conducted according to the preferred reporting items for systematic review and meta-analyses (PRISMA) guidelines (Moher et al. Citation2009).
Gene Ontology enrichment analysis
To overview the characteristics of genes, transcripts, and proteins obtained in the present study, we performed a GO analysis. We highlighted the most enriched biological processes, molecular functions, and cellular components from genomic (rare sequence variants), epigenomic, transcriptomic, and proteomic data for each sample type separately. The analysis was performed using the Enrichr tool (Xie et al. Citation2021). A Venn diagram of the data was made, using a web tool (Bioinformatics Citation2023). STRING was additionally used for retrieving protein interaction and functional information of the studied components (Szklarczyk et al. Citation2019).
Ethics approval
The data included in our review, retrieved from the PubMed repository was publicly available; therefore, no ethics permissions were required.
Authors’ contributions
Performed literature screening, curated and interpreted the data, and performed the enrichment analysis: RP. Provided technical advice for the article organization: AH and MS. Coordinated the study and revised the manuscript: TK and BP. Provided scientific advice from the clinical perspective: BP. All authors approved the final manuscript.
| Abbreviations | ||
| NGS | = | next-generation sequencing |
| WES | = | whole exome sequencing |
| WGS | = | whole genome sequencing |
| MMAF | = | multiple morphological abnormalities of the sperm flagella |
| GO | = | Gene Ontology |
| MS | = | mass spectrometry |
| DIGE | = | differential gel electrophoresis |
| PGT-M | = | preimplantation genetic testing for monogenic disorders |
Supplemental Material
Download MS Word (265.5 KB)Supplemental Material
Download MS Word (2.1 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Additional information
Funding
References
- Aderem A. 2005. Systems biology: its practice and challenges. Cell. 121(4):511–513. doi: 10.1016/j.cell.2005.04.020.
- Agarwal A, Mulgund A, Hamada A, Chyatte MR. 2015. A unique view on male infertility around the globe. Reprod Biol Endocrinol. 13(1):37. doi: 10.1186/s12958-015-0032-1.
- Bakey Z, Cabrera OA, Hoefele J, Antony D, Wu K, Stuck MW, Micha D, Eguether T, Smith AO, van der Wel NN, et al. 2023. IFT74 variants cause skeletal ciliopathy and motile cilia defects in mice and humans. PLoS Genet. 19(6):e1010796. doi: 10.1371/journal.pgen.1010796.
- Bansal SK, Gupta N, Sankhwar SN, Rajender S. 2015. Differential genes expression between fertile and infertile spermatozoa revealed by transcriptome analysis. PLOS One. 10(5):e0127007. doi: 10.1371/journal.pone.0127007.
- Barišić A, Buretić Tomljanović A, Starčević Čizmarević N, Ostojić S, Romac P, Vraneković J. 2021. A rare Y-autosome translocation found in a patient with nonobstructive azoospermia: case report. Syst Biol Reprod Med. 67(4):307–313. doi: 10.1080/19396368.2021.1898701.
- Berkay EG, Karaman B, Başaran S. 2023. A rare ring chromosome 21 abnormality is associated with azoospermia in two different phenotypically normal cases. Syst Biol Reprod Med. 69(5):387–393. doi: 10.1080/19396368.2023.2225682.
- Bioinformatics. 2023. Calculate and draw custom Venn diagrams. Bioinformatics & evolutionary genomics; [accessed 12 Jan]. https://bioinformatics.psb.ugent.be/webtools/Venn/.
- Blaurock J, Baumann S, Grunewald S, Schiller J, Engel KM. 2022. Metabolomics of human semen: a review of different analytical methods to unravel biomarkers for male fertility disorders. Int J Mol Sci. 23(16):9031. doi: 10.3390/ijms23169031.
- Coutton C, Vargas AS, Amiri-Yekta A, Kherraf ZE, Ben Mustapha SF, Le Tanno P, Wambergue-Legrand C, Karaouzène T, Martinez G, Crouzy S, et al. 2018. Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat Commun. 9(1):686. doi: 10.1038/s41467-017-02792-7.
- Dai X, Shen L. 2022. Advances and trends in omics technology development. Front Med. 9:911861. doi: 10.3389/fmed.2022.911861.
- Danshina PV, Geyer CB, Dai Q, Goulding EH, Willis WD, Kitto GB, McCarrey JR, Eddy EM, O'Brien DA. 2010. Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biol Reprod. 82(1):136–145. doi: 10.1095/biolreprod.109.079699.
- Di Palma T, Filippone MG, Pierantoni GM, Fusco A, Soddu S, Zannini M. 2013. Pax8 has a critical role in epithelial cell survival and proliferation. Cell Death Dis. 4(7):e729. doi: 10.1038/cddis.2013.262.
- Djureinovic D, Fagerberg L, Hallström B, Danielsson A, Lindskog C, Uhlén M, Pontén F. 2014. The human testis-specific proteome defined by transcriptomics and antibody-based profiling. Mol Hum Reprod. 20(6):476–488. doi: 10.1093/molehr/gau018.
- Dohle GR, Colpi GM, Hargreave TB, Papp GK, Jungwirth A, Weidner W, EAU Working Group on Male Infertility. 2005. EAU guidelines on male infertility. Eur Urol. 48(5):703–711. doi: 10.1016/j.eururo.2005.06.002.
- Edelbrock MA, Kaliyaperumal S, Williams KJ. 2013. Structural, molecular and cellular functions of MSH2 and MSH6 during DNA mismatch repair, damage signaling and other noncanonical activities. Mutat Res. 743–744:53–66. doi: 10.1016/j.mrfmmm.2012.12.008.
- Fainberg J, Hayden RP, Schlegel PN. 2019. Fertility management of Klinefelter syndrome. Expert Rev Endocrinol Metab. 14(6):369–380. doi: 10.1080/17446651.2019.1671821.
- Gao Y, Wu H, Xu Y, Shen Q, Xu C, Geng H, Lv M, Tan Q, Li K, Tang D, et al. 2021. Novel biallelic mutations in SLC26A8 cause severe asthenozoospermia in humans owing to midpiece defects: insights into a putative dominant genetic disease. Hum Mutat. 43(3):434–443. doi: 10.1002/humu.24322.
- Garrido N, Hervás I. 2020. Personalized medicine in infertile men. Urol Clin North Am. 47(2):245–255. doi: 10.1016/j.ucl.2019.12.011.
- Gupta N, Sarkar S, Mehta P, Sankhwar SN, Rajender S. 2022. Polymorphisms in the HSF2, LRRC6, MEIG1 and PTIP genes correlate with sperm motility in idiopathic infertility. Andrologia. 54(9):e14517. doi: 10.1111/and.14517.
- Hall EA, Keighren M, Ford MJ, Davey T, Jarman AP, Smith LB, Jackson IJ, Mill P. 2013. Acute versus chronic loss of mammalian Azi1/Cep131 results in distinct ciliary phenotypes. PLoS Genet. 9(12):e1003928. doi: 10.1371/journal.pgen.1003928.
- Hasin Y, Seldin M, Lusis A. 2017. Multi-omics approaches to disease. Genome Biol. 18(1):83. doi: 10.1186/s13059-017-1215-1.
- Hodžić A, Maver A, Plaseska-Karanfilska D, Ristanović M, Noveski P, Zorn B, Terzic M, Kunej T, Peterlin B. 2021. De novo mutations in idiopathic male infertility—a pilot study. Andrology. 9(1):212–220. doi: 10.1111/andr.12897.
- Houston BJ, Riera-Escamilla A, Wyrwoll MJ, Salas-Huetos A, Xavier MJ, Nagirnaja L, Friedrich C, Conrad DF, Aston KI, Krausz C, et al. 2021. A systematic review of the validated monogenic causes of human male infertility: 2020 update and a discussion of emerging gene–disease relationships. Hum Reprod Update. 28(1):15–29. doi: 10.1093/humupd/dmab030.
- Hunter N, Borts RH. 1997. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 11(12):1573–1582. doi: 10.1101/gad.11.12.1573.
- Ito C, Makino T, Mutoh T, Kikkawa M, Toshimori K. 2023. The association of ODF4 with AK1 and AK2 in mice is essential for fertility through its contribution to flagellar shape. Sci Rep. 13(1):2969. doi: 10.1038/s41598-023-28177-z.
- Jaillard S, McElreavy K, Robevska G, Akloul L, Ghieh F, Sreenivasan R, Beaumont M, Bashamboo A, Bignon-Topalovic J, Neyroud AS, et al. 2020. STAG3 homozygous missense variant causes primary ovarian insufficiency and male non-obstructive azoospermia. Mol Hum Reprod. 26(9):665–677. doi: 10.1093/molehr/gaaa050.
- Jungwirth A, Giwercman A, Tournaye H, Diemer T, Kopa Z, Dohle G, Krausz C, European Association of Urology Working Group on Male Infertility. 2012. European Association of Urology guidelines on Male Infertility: the 2012 update. Eur Urol. 62(2):324–332. doi: 10.1016/j.eururo.2012.04.048.
- Khazaie Y, Nasr Esfahani MH. 2014. MicroRNA and male infertility: a potential for diagnosis. Int J Fertil Steril. 8(2):113–118.
- Konno A, Ikegami K, Konishi Y, Yang HJ, Abe M, Yamazaki M, Sakimura K, Yao I, Shiba K, Inaba K, et al. 2016. Ttll9–/– mice sperm flagella show shortening of doublet 7, reduction of doublet 5 polyglutamylation and a stall in beating. J Cell Sci. 129(14):2757–2766. doi: 10.1242/jcs.185983.
- Krausz C, Escamilla AR, Chianese C. 2015. Genetics of male infertility: from research to clinic. Reproduction. 150(5):R159–R174. doi: 10.1530/REP-15-0261.
- Krzastek SC, Farhi J, Gray M, Smith RP. 2020. Impact of environmental toxin exposure on male fertility potential. Transl Androl Urol. 9(6):2797–2813. doi: 10.21037/tau-20-685.
- Krzastek SC, Smith RP, Kovac JR. 2020. Future diagnostics in male infertility: genomics, epigenetics, metabolomics and proteomics. Transl Androl Urol. 9(Suppl. 2):S195–S205. doi: 10.21037/tau.2019.10.20.
- Kukurba KR, Montgomery SB. 2015. RNA sequencing and analysis. Cold Spring Harb Protoc. 2015(11):951–969. doi: 10.1101/pdb.top084970.
- Leaver RB. 2016. Male infertility: an overview of causes and treatment options. Br J Nurs. 25(18):S35–S40. doi: 10.12968/bjon.2016.25.18.S35.
- Lee VCY, Chow JFC, Yeung WSB, Ho PC. 2017. Preimplantation genetic diagnosis for monogenic diseases. Best Pract Res Clin Obstet Gynaecol. 44:68–75. doi: 10.1016/j.bpobgyn.2017.04.001.
- Li P, Ji Z, Zhi E, Zhang Y, Han S, Zhao L, Tian R, Chen H, Huang Y, Zhang J, et al. 2022. Novel bi-allelic MSH4 variants causes meiotic arrest and non-obstructive azoospermia. Reprod Biol Endocrinol. 20(1):21. doi: 10.1186/s12958-022-00900-x.
- Li W, Tang W, Teves ME, Zhang Z, Zhang L, Li H, Archer KJ, Peterson DL, Williams DC Jr, Strauss JF 3rd, et al. 2015. A MEIG1/PACRG complex in the manchette is essential for building the sperm flagella. Development. 142(5):921–930. doi: 10.1242/dev.119834.
- Li Y, Wang Y, Wen Y, Zhang T, Wang X, Jiang C, Zheng R, Zhou F, Chen D, Yang Y, et al. 2021. Whole-exome sequencing of a cohort of infertile men reveals novel causative genes in teratozoospermia that are chiefly related to sperm head defects. Hum Reprod. 37(1):152–177. doi: 10.1093/humrep/deab229.
- Liu J, Rahim F, Zhou J, Fan S, Jiang H, Yu C, Chen J, Xu J, Yang G, Shah W, et al. 2023. Loss-of-function variants in KCTD19 cause non-obstructive azoospermia in humans. iScience. 26(7):107193. doi: 10.1016/j.isci.2023.107193.
- Liu W, He X, Yang S, Zouari R, Wang J, Wu H, Kherraf ZE, Liu C, Coutton C, Zhao R, et al. 2019. Bi-allelic mutations in TTC21A induce asthenoteratospermia in humans and mice. Am J Hum Genet. 104(4):738–748. doi: 10.1016/j.ajhg.2019.02.020.
- Liu X, Li Q, Wang W, Liu F. 2019. Aberrant expression of sperm‑specific glycolytic enzymes are associated with poor sperm quality. Mol Med Rep. 19(4):2471–2478. doi: 10.3892/mmr.2019.9926.
- Liu XG, Hu HY, Guo YH, Sun YP. 2016. Correlation between Y chromosome microdeletion and male infertility. Genet Mol Res. 15(2). doi: 10.4238/gmr.15028426.
- Llavanera M, Delgado-Bermúdez A, Ribas-Maynou J, Salas-Huetos A, Yeste M. 2022. A systematic review identifying fertility biomarkers in semen: a clinical approach through omics to diagnose male infertility. Fertil Steril. 118(2):291–313. doi: 10.1016/j.fertnstert.2022.04.028.
- Lundy SD, Sangwan N, Parekh NV, Selvam MKP, Gupta S, McCaffrey P, Bessoff K, Vala A, Agarwal A, Sabanegh ES, et al. 2021. Functional and taxonomic dysbiosis of the gut, urine, and semen microbiomes in male infertility. Eur Urol. 79(6):826–836. doi: 10.1016/j.eururo.2021.01.014.
- Lutzmann M, Grey C, Traver S, Ganier O, Maya-Mendoza A, Ranisavljevic N, Bernex F, Nishiyama A, Montel N, Gavois E, et al. 2012. MCM8- and MCM9-deficient mice reveal gametogenesis defects and genome instability due to impaired homologous recombination. Mol Cell. 47(4):523–534. doi: 10.1016/j.molcel.2012.05.048.
- Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK. 2013. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLOS One. 8(1):e55387. doi: 10.1371/journal.pone.0055387.
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. 2008. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18(9):1509–1517. doi: 10.1101/gr.079558.108.
- Martinez G, Coutton C, Loeuillet C, Cazin C, Muroňová J, Boguenet M, Lambert E, Dhellemmes M, Chevalier G, Hograindleur JP, et al. 2022. Oligogenic heterozygous inheritance of sperm abnormalities in mouse. Elife. 11:e75373. doi: 10.7554/eLife.75373.
- Mikec Š, Kolenc Ž, Peterlin B, Horvat S, Pogorevc N, Kunej T. 2022. Syndromic male subfertility: a network view of genome–phenome associations. Andrology. 10(4):720–732. doi: 10.1111/andr.13167.
- Mikeska T, Craig JM. 2014. DNA methylation biomarkers: cancer and beyond. Genes. 5(3):821–864. doi: 10.3390/genes5030821.
- Misra BB, Langefeld CD, Olivier M, Cox LA. 2018. Integrated omics: tools, advances, and future approaches. J Mol Endocrinol. doi: 10.1530/JME-18-0055.
- Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. 2009. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6(7):e1000097. doi: 10.1371/journal.pmed.1000097.
- Ning M, Lo EH. 2010. Opportunities and challenges in omics. Transl Stroke Res. 1(4):233–237. doi: 10.1007/s12975-010-0048-y.
- Omolaoye TS, Omolaoye VA, Kandasamy RK, Hachim MY, Du Plessis SS. 2022. Omics and Male Infertility: highlighting the application of transcriptomic data. Life. 12(2):280. doi: 10.3390/life12020280.
- Oud MS, Smits RM, Smith HE, Mastrorosa FK, Holt GS, Houston BJ, de Vries PF, Alobaidi BKS, Batty LE, Ismail H, et al. 2022. A de novo paradigm for male infertility. Nat Commun. 13(1):154. doi: 10.1038/s41467-021-27132-8.
- Peterlin A, Kunej T, Peterlin B. 2019. The role of circadian rhythm in male reproduction. Curr Opin Endocrinol Diabetes Obes. 26(6):313–316. doi: 10.1097/MED.0000000000000512.
- Pirih N, Kunej T. 2017. Toward a taxonomy for multi-omics science? Terminology development for whole genome study approaches by omics technology and hierarchy. OMICS. 21(1):1–16. doi: 10.1089/omi.2016.0144.
- Punab M, Poolamets O, Paju P, Vihljajev V, Pomm K, Ladva R, Korrovits P, Laan M. 2017. Causes of male infertility: a 9-year prospective monocentre study on 1737 patients with reduced total sperm counts. Hum Reprod. 32(1):18–31. doi: 10.1093/humrep/dew284.
- Rastgar Rezaei Y, Zarezadeh R, Nikanfar S, Oghbaei H, Nazdikbin N, Bahrami-Asl Z, Zarghami N, Ahmadi Y, Fattahi A, Nouri M, et al. 2021. microRNAs in the pathogenesis of non-obstructive azoospermia: the underlying mechanisms and therapeutic potentials. Syst Biol Reprod Med. 67(5):337–353. doi: 10.1080/19396368.2021.1951890.
- Seal RL, Braschi B, Gray K, Jones TEM, Tweedie S, Haim-Vilmovsky L, Bruford EA. 2023. Genenames.org: the HGNC resources in 2023. Nucleic Acids Res. 51(D1):D1003–D1009. doi: 10.1093/nar/gkac888.
- Seal RL, Chen LL, Griffiths-Jones S, Lowe TM, Mathews MB, O'Reilly D, Pierce AJ, Stadler PF, Ulitsky I, Wolin SL, et al. 2020. A guide to naming human non-coding RNA genes. EMBO J. 39(6):e103777. doi: 10.15252/embj.2019103777.
- Sha Y, Liu W, Nie H, Han L, Ma C, Zhang X, Xiao Z, Qin W, Jiang X, Wei X. 2022. Homozygous mutation in DNALI1 leads to asthenoteratozoospermia by affecting the inner dynein arms. Front Endocrinol. 13:1058651. doi: 10.3389/fendo.2022.1058651.
- Sha YW, Sha YK, Ji ZY, Mei LB, Ding L, Zhang Q, Qiu PP, Lin SB, Wang X, Li P, et al. 2018. TSGA10 is a novel candidate gene associated with acephalic spermatozoa. Clin Genet. 93(4):776–783. doi: 10.1111/cge.13140.
- Shen Y, Zhang F, Li F, Jiang X, Yang Y, Li X, Li W, Wang X, Cheng J, Liu M, et al. 2019. Loss-of-function mutations in QRICH2 cause male infertility with multiple morphological abnormalities of the sperm flagella. Nat Commun. 10(1):433. doi: 10.1038/s41467-018-08182-x.
- Silber SJ, Repping S. 2002. Transmission of male infertility to future generations: lessons from the Y chromosome. Hum Reprod Update. 8(3):217–229. doi: 10.1093/humupd/8.3.217.
- Sobotka V, Vozdova M, Heracek J, Rubes J. 2015. A rare Robertsonian translocation rob(14;22) carrier with azoospermia, meiotic defects, and testicular sperm aneuploidy. Syst Biol Reprod Med. 61(4):245–250. doi: 10.3109/19396368.2015.1045089.
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47(D1):D607–D613. doi: 10.1093/nar/gky1131.
- Tenenbaum-Rakover Y, Weinberg-Shukron A, Renbaum P, Lobel O, Eideh H, Gulsuner S, Dahary D, Abu-Rayyan A, Kanaan M, Levy-Lahad E, et al. 2015. Minichromosome maintenance complex component 8 (MCM8) gene mutations result in primary gonadal failure. J Med Genet. 52(6):391–399. doi: 10.1136/jmedgenet-2014-102921.
- Tu C, Nie H, Meng L, Yuan S, He W, Luo A, Li H, Li W, Du J, Lu G, et al. 2019. Identification of DNAH6 mutations in infertile men with multiple morphological abnormalities of the sperm flagella. Sci Rep. 9(1):15864. doi: 10.1038/s41598-019-52436-7.
- Wagner AO, Turk A, Kunej T. 2023. Towards a multi-omics of male infertility. World J Mens Health. 41(2):272–288. doi: 10.5534/wjmh.220186.
- Wang M, Yang QY, Zhou JP, Tan HP, Hu J, Jin L, Zhu LX. 2023. Novel compound heterozygous mutations in DNAH1 cause primary infertility in Han Chinese males with multiple morphological abnormalities of the sperm flagella. Asian J Androl. 25(4):512–519. doi: 10.4103/aja202292.
- Wei X, Sha Y, Wei Z, Zhu X, He F, Zhang X, Liu W, Wang Y, Lu Z. 2021. Bi-allelic mutations in DNAH7 cause asthenozoospermia by impairing the integrality of axoneme structure. Acta Biochim Biophys Sin. 53(10):1300–1309. doi: 10.1093/abbs/gmab113.
- Wistuba J, Mittag J, Luetjens CM, Cooper TG, Yeung CH, Nieschlag E, Bauer K. 2007. Male congenital hypothyroid 2007. Pax8–/– mice are infertile despite adequate treatment with thyroid hormone. J Endocrinol. 192(1):99–109. doi: 10.1677/JOE-06-0054.
- Wu H, Liu Y, Li Y, Li K, Xu C, Gao Y, Lv M, Guo R, Xu Y, Zhou P, et al. 2023. DNALI1 deficiency causes male infertility with severe asthenozoospermia in humans and mice by disrupting the assembly of the flagellar inner dynein arms and fibrous sheath. Cell Death Dis. 14(2):127. doi: 10.1038/s41419-023-05653-y.
- Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, et al. 2021. Gene set knowledge discovery with Enrichr. Curr Protoc. 1(3):e90. doi: 10.1002/cpz1.90.
- Xu X, Sha YW, Mei LB, Ji ZY, Qiu PP, Ji H, Li P, Wang T, Li L. 2018. A familial study of twins with severe asthenozoospermia identified a homozygous SPAG17 mutation by whole-exome sequencing. Clin Genet. 93(2):345–349. doi: 10.1111/cge.13059.
- Yap YT, Li W, Huang Q, Zhou Q, Zhang D, Sheng Y, Mladenovic-Lucas L, Yee SP, Orwig KE, Granneman JG, et al. 2023. DNALI1 interacts with the MEIG1/PACRG complex within the manchette and is required for proper sperm flagellum assembly in mice. Elife. 12:e79620. doi: 10.7554/eLife.79620.
- Zegers-Hochschild F, Adamson GD, de Mouzon J, Ishihara O, Mansour R, Nygren K, Sullivan E, Vanderpoel S, International Committee for Monitoring Assisted Reproductive Technology; World Health Organization. 2009. International Committee for Monitoring Assisted Reproductive Technology (ICMART) and the World Health Organization (WHO) revised glossary of ART terminology, 2009. Fertil Steril. 92(5):1520–1524. doi: 10.1016/j.fertnstert.2009.09.009.
- Zhang J, Mu X, Xia Y, Martin FL, Hang W, Liu L, Tian M, Huang Q, Shen H. 2014. Metabolomic analysis reveals a unique urinary pattern in normozoospermic infertile men. J Proteome Res. 13(6):3088–3099. doi: 10.1021/pr5003142.
- Zhang Z, Shen X, Gude DR, Wilkinson BM, Justice MJ, Flickinger CJ, Herr JC, Eddy EM, Strauss JF 3rd. 2009. MEIG1 is essential for spermiogenesis in mice. Proc Natl Acad Sci U S A. 106(40):17055–17060. doi: 10.1073/pnas.0906414106.
- Zupanic A, Bernstein HC, Heiland I. 2020. Systems biology: current status and challenges. Cell Mol Life Sci. 77(3):379–380. doi: 10.1007/s00018-019-03410-z.