ABSTRACT
Senescence frequently occurs in cancer cells in response to chemotherapy (called therapy-induced senescence). Senescent cells can exert paracrine effects through the senescence-associated secretory phenotype (SASP) promoting cancer recurrence and chemoresistance. The altered gut microbiota has been closely associated with cancer progression through the direct interaction with cancer cells. However, little is known about the relationship between the gut microbiota and therapy-induced senescent cells. This study aimed to explore the impact of the gut microbiota on therapy-induced senescent cells and the SASP. We found that esophageal squamous cell carcinoma (ESCC) cells were induced into senescence following platinum-based chemotherapy, accompanied by the secretion of a robust SASP. Furthermore, senescent ESCC cells exerted a tumor-promoting effect through the SASP both in vitro and in vivo. Through 16S rRNA gene sequencing and fluorescence in situ hybridization, we identified that Fusobacterium nucleatum (F. nucleatum) was abundant in human ESCC cancerous tissues and correlated with poor prognosis in ESCC patients. Notably, F. nucleatum further promoted the secretion of the SASP by senescent ESCC cells. Compared with the conditioned medium from senescent ESCC cells, the conditioned medium from F. nucleatum-treated senescent ESCC cells accelerated tumor growth in xenograft models, enhanced migration and invasion abilities, and potentiated chemoresistance both in vitro and in vivo. Mechanistically, F. nucleatum invaded and survived in senescent ESCC cells and induced an increase in DNA damage to further activate the DNA damage response pathway, thus enhancing the SASP. Altogether, these findings reveal for the first time that F. nucleatum promotes the secretion of chemotherapy-induced SASP to drive ESCC progression and chemoresistance, which supports F. nucleatum as a potential target for ESCC therapy.
1. Introduction
Esophageal cancer is the seventh most common malignant cancer worldwide and the sixth leading cause of cancer-related deaths with the 5-year survival rate of 15%-25%.Citation1 Esophageal squamous cell carcinoma (ESCC) is a histologic subtype constituting more than 90% of all esophageal cancer cases.Citation2 Platinum-based chemotherapy such as cisplatin (CDDP) is widely used for the first-line treatment of patients with ESCC.Citation3 However, ESCC patients frequently experience chemoresistance and cancer relapse after treatment with platinum-based chemotherapy.Citation3 Thus, there is an urgent need to clarify the underlying mechanisms and develop new and improved treatment strategies.
Cellular senescence is a steady state of cell growth arrest that is evoked by a wide range of intrinsic and extrinsic stimuli, including telomere shortening, oncogenic activation, oxidative and genotoxic stress, and mitochondrial dysfunction.Citation4 It is now well established that cancer cells can be induced into senescence during several cancer interventions such as chemotherapy and radiotherapy.Citation5,Citation6 Unlike apoptotic cells, senescent cells do not die immediately and gradually accumulate in the body following cancer treatment. Moreover, senescent cells remain metabolically active and secrete a plethora of proinflammatory factors including cytokines, chemokines, growth factors, and matrix metalloproteinases, which are collectively termed the senescence-associated secretory phenotype (SASP).Citation7 Given the stimulatory effects of the SASP on neighboring cells through paracrine actions, accumulating evidence suggests that therapy-induced senescent cells and the SASP are closely associated with the outcomes of cancer treatment.Citation8–10 For example, CDDP-induced cellular senescence in ovarian cancer contributed to chemoresistance through the induction of senescence-associated cancer stem cells by promoting the SASP.Citation11 CDDP-induced senescent melanoma cells accelerated neighboring non-senescent melanoma cell growth through the paracrine actions of the SASP, leading to cancer recurrence.Citation12 Additionally, in an orthotopic xenograft model of papillary thyroid carcinoma, senescent cancer cells were able to promote the invasion and migration of cancer cells through the SASP.Citation13 Of note, recent in vivo studies have shown that the selective elimination of chemotherapy-induced senescent cells significantly reduced cancer relapse.Citation14,Citation15 Therefore, therapy-induced senescent cells and the SASP are being considered as one of the most crucial contributors to chemoresistance and cancer recurrence.
In recent years, increasing evidence has suggested that the gut microbiota is closely linked to the development, progression and prognosis of multiple cancers.Citation16 Fusobacterium nucleatum (F. nucleatum), an anaerobic Gram-negative bacterium found in the human oral and gastrointestinal tract, attracts most attention due to its important contributions to human cancers.Citation17 Recent studies demonstrated that F. nucleatum promoted cancer cell growth, migration, and invasion through activation of various cancer-promoting signaling pathways, thereby facilitating the initiation and progression of gastrointestinal cancers.Citation18–20 Furthermore, F. nucleatum was reported to confer chemoresistance in cancer cells via the induction of autophagy.Citation21,Citation22 Notably, the majority of current studies mainly focus on the direct and indirect effects of F. nucleatum on cancer cells, while with few on its interaction with therapy-induced senescent cells. Because senescent cell production often occurs during cancer therapy, it is of great importance to ascertain whether F. nucleatum can affect therapy-induced senescent cells and the secretion of the SASP to provoke cancer relapse and chemoresistance, which will provide us a more comprehensive understanding of the pro-tumorigenic function of F. nucleatum.
In this study, we found for the first time that F. nucleatum could invade and survive in chemotherapy-induced senescent ESCC cells and promote the secretion of the SASP, thereby contributing to ESCC progression and chemoresistance. Our findings provide a novel insight into the mechanisms by which F. nucleatum drives the development of ESCC, and further support F. nucleatum as a potential target for ESCC treatment.
2. Results
2.1 CDDP induced senescence and a robust SASP in p53 wild-type human ESCC cell lines and in human ESCC tissues
Considering the critical role of p53 in the induction of cellular senescence, two p53 wild-type human ESCC cell lines KYSE-150 and ECA-109 were utilized in our experiments. To evaluate whether CDDP could induce senescence in ESCC cells in vitro, ESCC cells were treated with various concentrations of CDDP for 96 h. CCK-8 assay showed that CDDP significantly decreased cell viability of ESCC cells in a concentration-dependent manner (Figure S1a). We observed that nearly 90% of CDDP-treated ESCC cells (KYSE-150 treated with 0.5 μg/ml CDDP and ECA-109 with 1 μg/ml CDDP) displayed an enlarged and flattened morphological alteration, thus implying cell senescence. Next, the activity of senescence-associated beta-galactosidase (SA-β-gal), a canonical marker of cell senescence was measured.Citation23 CDDP remarkably increased the percentage of SA-β-gal-positive cells (). Of note, this concentration of CDDP potently inhibited the growth of ESCC cells (Figure S1b) and only caused slight toxicity as verified by weak up-regulation of two widely used biomarkers of cellular apoptosis, cleaved PARP and cleaved Caspase-3 (Figure S1c). In the following experiments in vitro, 0.5 μg/ml CDDP used in KYSE-150 cells and 1 μg/ml CDDP in ECA-109 cells were set as default unless otherwise indicated.
Figure 1. CDDP induced senescence and a robust SASP in p53 wild-type ESCC cells and in human ESCC cancerous tissues.
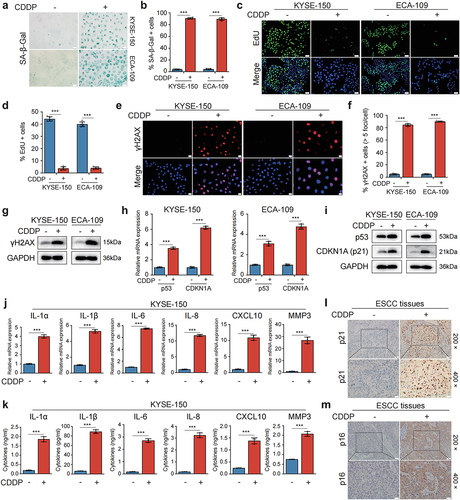
To validate the results obtained by SA-β-gal staining, we next monitored cell proliferation by EdU incorporation assay. The fraction of EdU-positive cells was dramatically reduced with CDDP treatment compared to control group, indicating the impaired proliferation activity of CDDP-treated ESCC cells (). Furthermore, we also performed colony formation assay to analyze cell proliferation. A dramatic loss in colony formation was observed in ESCC cells after treatment with CDDP (Figure S1d). In addition to the stagnation of cell growth, another representative characteristic of senescent cells is their stable cell cycle arrest, we therefore analyzed cell cycle distribution of CDDP-treated ESCC cells. Cell cycle analysis by flow cytometry demonstrated that more than 80% of ESCC cells were halted at G2/M phase upon treatment with CDDP for 96 h, accompanied by a marked decrease in the proportion of G1 and S phases (Figure S1e,f). Since the DNA damage response plays an important role in the induction of cellular senescence, we next examined a canonical DNA damage marker γH2AX after CDDP treatment. As expected, CDDP-treated cells had a remarkable increase in DNA damage over control cells (). Subsequently, the senescence-associated p53/p21 pathway mediated by the DNA damage response was also monitored. The elevated mRNA and protein expression of p53 and p21 (encoded by the CDKN1A gene) were observed in CDDP-treated ESCC cells (). Additionally, one of the most profound characteristics of senescence cells is the SASP, we thus detected the expression of several SASP-related factors that were frequently up-regulated in other senescent cells. The results of RT-qPCR and western blot demonstrated that after treatment with CDDP, the mRNA and protein expression of IL-1α, IL-1β, IL-6, and IL-8 were markedly increased ( and Figure S1g,h). However, up-regulation of CXCL10 and MMP3 was just observed in CDDP-treated KYSE-150 cells ( and Figure S1h), not in ECA-109 cells (Figure S1g,h), suggesting the composition of the SASP varied related to cell type undergoing senescence. The ELISA was used to determine the levels of these SASP-related factors in the conditioned medium (CM) from ESCC cells. The levels of these factors were significantly elevated in the CM from CDDP-treated ESCC cells compared with that in the CM from CDDP-untreated cells ( and Figure S1l). Together, these results indicated that CDDP was capable of inducing senescence and a robust SASP in p53 wild-type ESCC cell lines.
To further confirm whether CDDP could induce cancer cell senescence in human ESCC tissues, tumor specimen sections from ESCC patients who had received CDDP-based neoadjuvant chemotherapy prior to surgical resection were stained for senescence biomarkers p21 and p16 using immunohistochemical staining. Tumor sections from ESCC patients who had not been treated with chemotherapy before surgical resection were stained as controls. We observed that the rate of p21-positive staining in tumor sections treated with chemotherapy was significantly higher than that in the untreated tumor sections (p = 0.002, and ) as calculated using Pearson chi-square test. Moreover, the rate of p16-positive staining in the treated tumor sections was also higher than that in the untreated tumor sections (p = 0.012, and ). These results suggested that CDDP-based chemotherapy was able to induce cancer cell senescence in human ESCC tissues.
Table 1. The expression of p21 and p16 in cancerous tissues of ESCC patients treated with or without CDDP-based neoadjuvant chemotherapy.
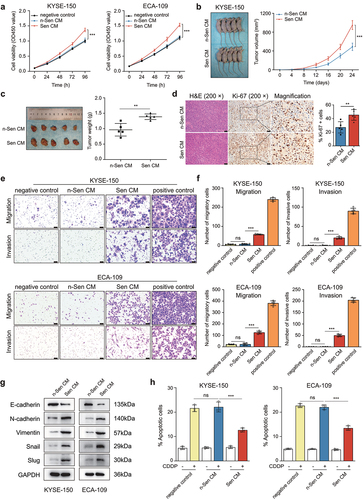
2.2 CDDP-induced SASP promoted tumorigenesis in the xenograft model and ESCC cell proliferation, migration, invasion, and chemoresistance
Previous studies showed that the SASP secreted by senescent cells could promote the malignant behaviors of non-senescent cancer cells,Citation24 we therefore investigated the effect of CDDP-induced SASP on ESCC cell growth in vitro. First, the CM from CDDP-treated ESCC cells (defined as Sen CM) and the CM from CDDP-untreated cells (as n-Sen CM) were collected and used for the culture of ESCC cells. The growth of Sen CM-treated ESCC cells was markedly increased when compared to that of n-Sen CM-treated cells, whereas there were no significant differences in the growth curves between RPMI-1640 medium-treated (defined as negative control) and n-Sen CM-treated ESCC cells (). Next, tumor xenograft experiments were performed to verify the role of CDDP-induced SASP in ESCC cells in vivo. KYSE-150 cells were subcutaneously inoculated into BALB/c nude mice. The mice were subsequently injected with Sen CM and n-Sen CM according to the experimental design plan. In comparison with n-Sen CM-treated group, Sen CM-treated group showed accelerated tumor growth and an increased tumor weight (). An enhanced staining of the proliferation marker Ki-67 was also observed in Sen CM-treated xenograft tissues (). Furthermore, we also analyzed the impact of CDDP-induced SASP on the migration and invasion ability of ESCC cells. The transwell assay demonstrated that Sen CM treatment significantly enhanced ESCC cell migration and invasion compared with the n-Sen CM-treated group, whereas there were no differences between the n-Sen CM-treated group and the negative control group (). In agreement with this, ESCC cells cultured with Sen CM displayed reduced protein expression of epithelial marker E-cadherin, elevated expression of mesenchymal markers N-cadherin and Vimentin, and up-regulation of epithelial-mesenchymal transition transcription factors Snail and Slug when compared with ESCC cells cultured with n-Sen CM, suggesting the promotional effect of CDDP-induced SASP on ESCC cell migration and invasion (). In addition, cell apoptosis was also assessed by flow cytometry after double staining with FITC-Annexin V/PI. The results showed that Sen CM treatment remarkably reduced the proportion of apoptotic cell death induced by CDDP compared with the n-Sen CM-treated group, whereas there were no apoptotic rate differences between the negative control group and the n-Sen CM-treated group ( and Figure S2a). Collectively, these in vitro and in vivo findings suggested that CDDP-induced senescent cells promoted the progression and chemoresistance of ESCC through the paracrine actions of the SASP.
Figure 2. CDDP-induced SASP promoted tumorigenesis in the xenograft model and ESCC cell growth, migration, invasion, and chemoresistance.
2.3 Gut microbiota composition was altered in ESCC tissues and F. nucleatum was correlated with poor prognosis and chemoresistance in ESCC patients
Recent studies have identified that the gut microbiota plays an important role in the development and prognosis of human cancers. To investigate the gut microbiota profile in ESCC patients, we carried out 16S rRNA gene sequencing to analyze the microbiome in cancerous and paired adjacent normal tissues of resected ESCC samples. We first analyzed the differences in the microbial α diversity between the Cancer group and the Normal group using the different α diversity indexes. The Simpson and Shannon indexes were applied to determine the community diversity, and the Chao 1 and Ace indexes were used to evaluate the community richness. Significant differences were observed in the Simpson and Shannon indexes between the Cancer and Normal groups (), although there were no significant differences in the Chao 1 and Ace indexes between the two groups (Figure S3a,b). Then we compared the differences in the β diversity between the two groups via principal coordinate analysis and non-metric multidimensional scaling analysis. The results showed that there were significant differences in the β diversity between the two groups (Figure S3c,d). We further analyzed the specific composition and abundance differences of the microbiota community in the two groups. At the phylum level, Firmicutes, Bacteroidota, and Fusobacteriota were more abundant in the Cancer group than in the Normal group, whereas Proteobacteria and Actinobacteriota were less abundant ( and Figure S3e). At the genus level, the relative abundance of Peptostreptococcus, Porphyromonas, Prevotella, Alloprevotella, and Fusobacterium was higher and that of Pseudomonas, Rhodococcus, and Achromobacter was lower in the Cancer group than in the Normal group. ( and Figure S3f). Taken together, these results revealed the notable alteration in the composition of the gut microbiota in cancerous and adjacent normal tissues of ESCC patients.
Figure 3. Gut microbiota composition was altered in ESCC tissues and F. nucleatum was correlated with poor prognosis and chemoresistance in ESCC patients.
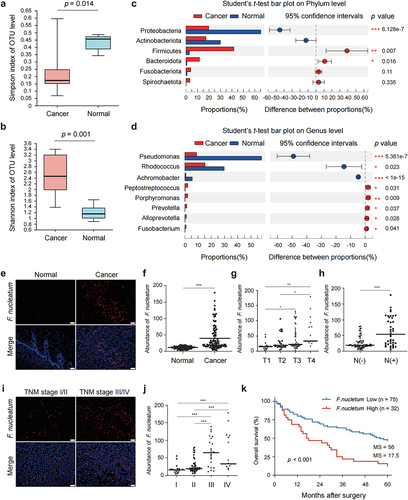
Of the Fusobacterium species, F. nucleatum is one of the most common members and has been closely implicated in the development, progression and prognosis of multiple gastrointestinal cancers such as colorectal cancer and pancreatic cancer. In the present study, we explored the role of F. nucleatum in ESCC progression, prognosis and chemoresistance. We first conducted fluorescence in situ hybridization (FISH) with a Cy3-labeled probe to detect F. nucleatum in the sections of formalin-fixed and paraffin-embedded cancerous and matched adjacent normal tissues of ESCC patients. The results of FISH showed that F. nucleatum was abundant in cancerous tissues compared to adjacent normal tissues. (). Notably, an interesting finding was that F. nucleatum was mainly distributed in clusters in cancerous tissues (). Furthermore, the abundance of F. nucleatum in cancerous tissues was positively correlated with tumor infiltration depth () and lymph node metastasis (). A higher abundance of F. nucleatum was also found in cancerous tissues from ESCC patients with TNM stage III and IV than those with TNM stage I and II (). In addition, X-tile software was used to determine the optimal cutoff value of F. nucleatum abundance in cancerous tissues for survival analysis.Citation25 The optimal cutoff value of F. nucleatum abundance was 47. Thus, ESCC patients were divided into two groups, F. nucleatum-low abundance, which had 75 cases, and F. nucleatum-high abundance, which had 32 cases. The differences of clinicopathological features and prognosis were compared between the two groups. There were significant differences in lymph node metastasis and tumor TNM stage between the two groups, while no significant differences were observed in age, gender, tumor location, tumor size, differentiation, and tumor infiltration depth (). Kaplan-Meier analysis showed that ESCC patients with F. nucleatum-high abundance had a significantly shorter median time of overall survival compared with those with F. nucleatum-low abundance (p < 0.001, ). Finally, we evaluated the relationship between F. nucleatum and chemotherapy response in 25 ESCC patients who had received CDDP-based neoadjuvant chemotherapy prior to surgical resection. We observed a significantly lower chemotherapeutic response rate in patients with F. nucleatum-high abundance than those with F. nucleatum-low abundance (p = 0.017, Figure S3g) as calculated using Fisher’s exact test. These results indicated that the abundance of F. nucleatum was significantly increased in ESCC cancerous tissues and positively correlated with progression, poor prognosis and chemoresistance in ESCC patients.
Table 2. Clinicopathological features in F. nucleatum-low vs. F. nucleatum-high ESCC patients.
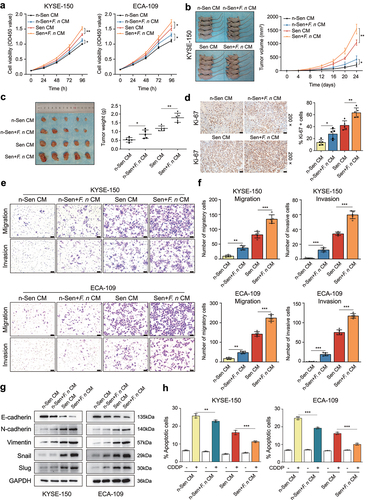
2.4 F. nucleatum infection exacerbated the SASP-driven malignant phenotypes and chemoresistance of ESCC cells in vitro and in vivo
Given the fact that senescent cells production often occurs during platinum-based chemotherapy for ESCC patients, it is important to reveal the underlying relationship between F. nucleatum and therapy-induced senescent ESCC cells. Therefore, we next investigated whether senescent cells after F. nucleatum stimulation further enhanced the SASP-driven ESCC cell proliferation, migration, invasion, and chemoresistance. First, in addition to Sen CM and n-Sen CM, the CM from F. nucleatum-treated senescent ESCC cells (defined as Sen+F. nucleatum CM) and the CM from F. nucleatum-treated non-senescent cells (as n-Sen+F. nucleatum CM) were harvested and utilized for ESCC cell culture. The growth rate of Sen+F. nucleatum CM-treated ESCC cells was significantly higher than that of Sen CM-treated cells. Notably, n-Sen+F. nucleatum CM treatment also promoted ESCC cell proliferation compared with n-Sen CM-treated group (). Next, we further assessed the stimulatory effect of senescent ESCC cells after F. nucleatum infection on ESCC cell growth in the xenograft model. KYSE-150 cells were subcutaneously inoculated into BALB/c nude mice. The mice were subsequently injected with Sen CM, Sen+F. nucleatum CM, n-Sen CM, and n-Sen+F. nucleatum CM according to the experimental design plan. As expected, Sen+F. nucleatum CM-treated group had a higher rate of tumor growth and a larger mean tumor weight compared to Sen CM-treated group. Meanwhile, n-Sen+F. nucleatum CM-treated group also showed accelerated tumor growth and an elevated tumor weight in comparison with n-Sen CM-treated group (). In agreement with this, the proportion of Ki-67-positive cells was significantly increased in Sen+F. nucleatum CM-treated xenograft tissues when compared with that in Sen CM-treated tissues, and there also was a remarked increase in the fraction of Ki-67-positive cells in n-Sen+F. nucleatum CM-treated group compared to n-Sen CM-treated group (). Furthermore, the transwell assay showed that Sen+F. nucleatum CM treatment dramatically promoted ESCC cell migration and invasion compared to the Sen CM-treated group, and there were significant differences between the n-Sen+F. nucleatum CM-treated group and the n-Sen CM-treated group, indicating the stimulatory effect of F. nucleatum infection on ESCC cell migration and invasion (). Consistent with these results, ESCC cells cultured with Sen+F. nucleatum CM also exhibited reduced E-cadherin expression, increased N-cadherin and Vimentin expression, and elevated expression of Snail and Slug compared with ESCC cells cultured with Sen CM (). The similar tendencies of these protein expression were also found in ESCC cells cultured with n-Sen+F. nucleatum CM in comparison with ESCC cells cultured with n-Sen CM (). Additionally, the results of apoptosis detected by flow cytometry showed that Sen+F. nucleatum CM treatment remarkably reduced the fraction of CDDP-induced cell apoptosis compared with the Sen CM-treated group ( and Figure S2b). Notably, n-Sen+F. nucleatum CM treatment also significantly decreased the apoptotic rates in comparison with n-Sen CM-treated group ( and Figure S2b). Together, these in vitro and in vivo observations demonstrated that F. nucleatum infection further aggravated the SASP-mediated ESCC progression and chemoresistance.
Figure 4. F. nucleatum stimulation aggravated the SASP-mediated malignant phenotypes and chemoresistance of ESCC cells.
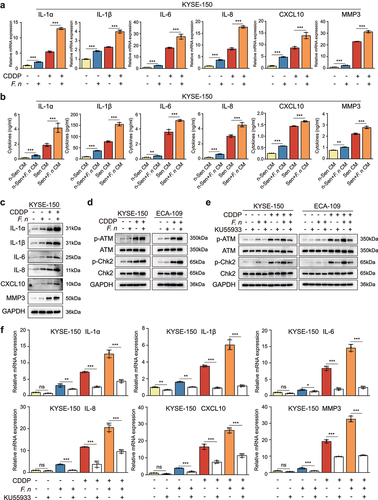
2.5 F. nucleatum infection enhanced the secretion of CDDP-induced SASP through activation of the DNA damage response pathway
To explore the mechanism by which F. nucleatum infection potentiated the SASP-driven ESCC progression and chemoresistance, the expression of SASP-related factors in ESCC cells after F. nucleatum stimulation was examined. The results of RT-qPCR and western blot showed that the mRNA and protein expression of SASP-related factors including IL-1α, IL-1β, IL-6, IL-8, CXCL10, and MMP3 were dramatically higher in F. nucleatum-infected senescent cells than in uninfected senescent cells ( and Figure S4a,b). Moreover, F. nucleatum stimulation alone also promoted the mRNA and protein expression of these factors ( and Figure S4a,b). The results of ELISA were consistent with these results ( and Figure S4c). Based on these findings, we confirmed that senescent cells after F. nucleatum infection further promoted ESCC progression and chemoresistance via enhancing the secretion of CDDP-induced SASP. Next, we investigated the underlying mechanism of the elevated expression of SASP-related factors after F. nucleatum infection. There were no differences in the fraction of senescent cells between the F. nucleatum-infected group and the uninfected group, suggesting the increased SASP expression by F. nucleatum infection was not related to the proportion of senescent cells (Figure S4d,e). Emerging evidence has revealed that F. nucleatum can regulate the expression of multiple cytokines through the TLR4/MyD88/NF-κB and E-cadherin/β-catenin signaling pathways. Citation19,Citation26 Thus, we tested the expression of TLR4, MyD88, p65, E-cadherin, and β-catenin. However, there were no changes in the expression of MyD88, p65, p-p65, E-cadherin, and β-catenin in ESCC cells after F. nucleatum infection (Figure S5a,b), although TLR4 was up-regulated as verified by RT-qPCR and western blot as well as immunofluorescent staining (Figure S5a-c), indicating the TLR4/MyD88/NF-κB and E-cadherin/β-catenin pathways were also not involved in the increased SASP expression by F. nucleatum infection. Interestingly, these two signaling pathways were remarkably activated in CDDP-induced senescent ESCC cells (Figure S5a,b). Previous studies have shown that the SASP can be orchestrated by multiple signaling pathways, in particular the DNA damage response pathway.Citation27 We therefore investigated whether F. nucleatum further promoted the CDDP-induced SASP via activation of the DNA damage response pathway. We measured the protein expression of DNA damage response-related molecules including p-ATM, ATM, p-Chk2, and Chk2 in ESCC cells after F. nucleatum infection. Western blot analysis demonstrated that in comparison with the uninfected group, the F. nucleatum-infected group exhibited the enhanced protein expression of p-ATM and p-Chk2, suggesting the further activation of the DNA damage response pathway in senescent ESCC cells after F. nucleatum infection (). To further confirm that F. nucleatum stimulation enhanced the secretion of CDDP-induced SASP via activating the DNA damage response pathway, we blocked the activation of the DNA damage response pathway by a selective ATM kinase inhibitor KU55933. As expected, the protein expression of p-ATM was significantly down-regulated by the treatment with KU55933, as was the level of its downstream molecule p-Chk2 (). More importantly, the mRNA levels of SASP-related factors including IL-1α, IL-1β, IL-6, IL-8, CXCL10, and MMP3 were markedly suppressed by KU55933 treatment ( and Figure S6a). In addition, there were no differences in the proportion of senescent cells before and after treatment with KU55933 (Figure S6b,c). Above all, these results confirmed that the enhanced secretion of CDDP-induced SASP caused by F. nucleatum infection was mainly mediated by the further activation of the DNA damage response pathway.
Figure 5. F. nucleatum infection enhanced the secretion of CDDP-induced SASP through the activation of DNA damage response pathway.
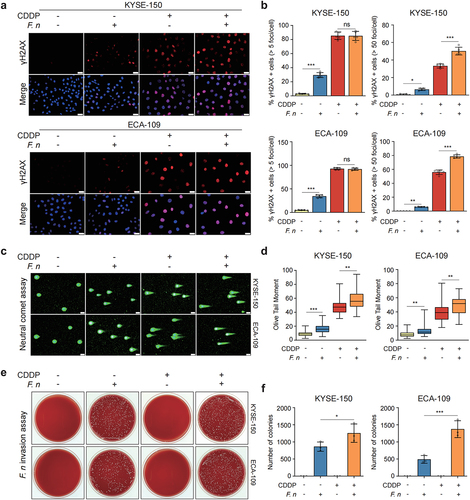
2.6 F. nucleatum invaded and survived in ESCC cells and induced DNA damage to activate the DNA damage response pathway
To further explore the mechanism by which F. nucleatum infection activated the DNA damage response pathway, we detected the levels of DNA damage in ESCC cells after F. nucleatum stimulation. The results of immunofluorescent staining demonstrated that the formation of γH2AX foci was remarkably increased in F. nucleatum-infected ESCC cells when compared to that in uninfected cells (). Interestingly, F. nucleatum stimulation aggravated DNA damage in senescent ESCC cells, but there was no further increase in the percentage of γH2AX-positive cells (). Moreover, western blot analysis also showed an increased DNA damage in ESCC cells after F. nucleatum infection (Figure S7a). Additionally, the neutral comet assay demonstrated that F. nucleatum infection significantly increased the value of olive tail moment in ESCC cells, indicating an increase in DNA damage of F. nucleatum-infected cells (). To determine the possible causes of F. nucleatum-induced DNA damage, we conducted F. nucleatum invasion assay. ESCC cells were co-cultured with F. nucleatum (MOI = 100) for 24 h and then incubated in the culture medium containing 200 μg/ml metronidazole and 300 μg/ml gentamicin for 1 h to completely kill extracellular F. nucleatum. To validate the effectiveness of this concentration of antibiotics on the removal of extracellular F. nucleatum, we plated the culture medium treated with antibiotics on the agar and cultured it under anaerobic condition for 72 h. We observed no colony formation on the agar (Figure S7b), indicating that extracellular F. nucleatum was completely eliminated by the antibiotics. Notably, we found that the cell lysate of F. nucleatum-infected ESCC cells formed colonies on the agar after 72 h under anaerobic conditions, which indicated that F. nucleatum invaded and survived in ESCC cells (). Furthermore, we observed that the number of colonies formed from F. nucleatum-infected senescent cells was more than that from F. nucleatum-infected non-senescent cells, suggesting that F. nucleatum was prone to invade senescent cells compared to non-senescent cells (). Together, these findings indicated that F. nucleatum invaded and survived in senescent ESCC cells and induced DNA damage to further activate the DNA damage response pathway, thereby promoting the secretion of CDDP-induced SASP.
Figure 6. F. nucleatum invaded and survived in ESCC cells and increased DNA damage.
3. Discussion
Cellular senescence was, in addition to apoptosis, initially considered to be another beneficial outcome of cancer therapy mainly due to senescence-mediated cell growth arrest.Citation28 However, emerging evidence has demonstrated that the accumulation of therapy-induced senescent cells contributes to cancer progression and therapeutic resistance through the paracrine actions of the SASP, which was described in detail in our previous review.Citation29 In this study, we observed that CDDP induced senescence and a robust SASP in human ESCC cell lines expressing wild-type p53. Furthermore, we confirmed the existence of CDDP-induced cancer cell senescence in cancerous tissues of ESCC patients. More importantly, CDDP-induced senescent cells promoted ESCC cell proliferation, migration, invasion, and chemoresistance via the SASP both in vitro and in vivo. Therefore, our current study provides the evidence that CDDP is able to induce ESCC cell senescence in vitro and in vivo, and also supports the idea that therapy-induced senescent cells are detrimental to the organism and thus should be eliminated in time after cancer therapy. Consistent with this, senotherapy emerged in recent years, which refers to selective removal of senescent cells, has been proposed as a promising adjuvant approach for the treatment of cancer, particularly in combination with currently used cancer therapy.Citation30–32 Many experiments have shown that the concurrent use of senotherapy and conventional cancer therapies significantly improves therapeutic efficacy and reduces the risk of cancer relapse.Citation14,Citation15,Citation33 Despite the potential use of senotherapy in ESCC treatment has not been reported, this is obviously worth further investigations. Notably, we used p53 wild-type ESCC cell lines to perform our present study because of the vital role of p53 in the induction of senescence. However, p53 is frequently mutated in human cancers.Citation34 Thus, it is necessary to further investigate whether CDDP can induce senescence in ESCC cells with mutant or deleted type p53 gene in the future studies. Additionally, due to the short follow-up period and the small number of patients included, the potential relationship between therapy-induced senescence and the prognosis of ESCC patients could not evaluate for the time being, but we will continue to expand the sample size, follow up these patients, and perform further statistical analyses in our future studies.
Emerging evidence has identified the close association between the gut microbiota and the development, progression and prognosis of multiple human cancers.Citation16 In our current study, by comparing microbial community profile in cancerous and adjacent normal tissues of ESCC patients using 16S rRNA gene sequencing, we found the significantly altered composition of the gut microbiota between the two groups and observed the remarkably elevated abundance of Fusobacterium species in cancerous tissues, which implied the potential role of Fusobacterium in ESCC progression. F. nucleatum, one of the most important members of the Fusobacterium species, has been found in esophageal cancer tissue and associated with patient prognosis.Citation35 Therefore, F. nucleatum was further detected using FISH in the sections of formalin-fixed and paraffin-embedded cancerous and adjacent normal tissues of ESCC patients. We observed that the abundance of F. nucleatum in cancerous tissues was significant higher than that in adjacent normal tissues. Furthermore, the abundance of F. nucleatum in cancerous tissues was closely associated with tumor infiltration depth, lymph node metastasis and tumor TNM stage. In addition, F. nucleatum in cancerous tissues contributed to chemoresistance and worse prognosis in ESCC patients.
In recent years, studies have reported some mechanisms on how F. nucleatum influences ESCC progression and chemoresistance. Nomoto et al. reported that F. nucleatum promoted the proliferation, migration, and invasion of ESCC cells through the activation of NOD1/NF-κB signaling pathway.Citation20 In our previous study, we showed that F. nucleatum accelerated ESCC cell proliferation via activating AHR/CYP1A1 signaling pathway.Citation36 In addition, high burden of F. nucleatum in ESCC tissues was correlated with poor response to chemotherapy in ESCC patients.Citation37 Liu et al. reported that F. nucleatum contributed to chemoresistance in ESCC cells via the induction of autophagy.Citation22 Liang et al. demonstrated recently that F. nucleatum increased the expression of NLRP3 followed by the enrichment of myeloid-derived suppressor cells, thereby leading to ESCC cell chemoresistance.Citation38 However, the underlying relationship between F. nucleatum and therapy-induced senescent ESCC cells to date remains unknown. In the present study, we observed that CDDP-induced senescent cells after co-cultured with F. nucleatum further exacerbated the SASP-driven ESCC cell proliferation, migration, invasion, and chemoresistance both in vitro and in vivo, suggesting the contribution of F. nucleatum to the secretion of the SASP by senescent cells. To the best of our knowledge, we are the first to establish a crosstalk between F. nucleatum and therapy-induced senescent cells and provide novel evidence for the pro-tumorigenic function of F. nucleatum in human cancers.
Available literature sources indicate that the SASP can be modulated by multiple signaling pathways including the DNA damage response pathway.Citation27,Citation39 In response to DNA damage caused by chemotherapeutic drugs, the DNA damage response pathway is activated to not only induce cell cycle arrest and more importantly promote the expression of various proinflammatory factors such as cytokines and chemokines. In our current study, we demonstrated that F. nucleatum further promoted the secretion of CDDP-induced SASP through the activation of the DNA damage response pathway. Moreover, F. nucleatum contributed to the activation of the DNA damage response pathway mainly due to an increased DNA damage in senescent and non-senescent cells. An interesting finding was that F. nucleatum invaded and survived in senescent and non-senescent cells, which may be a major cause of the increase in DNA damage. However, unlike other microorganisms such as colibactin-producing Escherichia coli can produce toxins that cause DNA damage,Citation40 F. nucleatum encodes no known toxins and very few canonical virulence factors.Citation41,Citation42 Recently, Fusobacterium adhesin A (FadA) was identified as an important virulence factor of F. nucleatum and verified to bind and invade host cells, thus provoking oncogenic and inflammatory responses.Citation26 A more recent study showed that FadA induced DNA damage and promoted genome instability, which facilitated cancer progression.Citation43 Therefore, we speculate that F. nucleatum activates the DNA damage response pathway in ESCC cells mainly due to DNA damage induced by FadA. However, further study is still needed to unravel the underlying mechanism of FadA-induced DNA damage. Additionally, we observed that F. nucleatum was prone to invade senescent cells compared to non-senescent cells. This may be due to: (i) a significant increase in the surface area of senescent cells to facilitate F. nucleatum invasion; (ii) the increased fragility of senescent cell membrane; (iii) the elevated expression of an adhesin receptor E-cadherin in senescent cells, which contributes to F. nucleatum invasion into senescent cells through interaction with FadA.Citation26
Given the critical role of the gut microbiota in defining chemotherapeutic efficacy, the gut microbes are undeniably potential candidates for being treatment targets. Antibiotics can remarkably alter the composition of the gut microbiota, which in turn influences the efficacy of chemotherapy, although the use of antibiotics is mainly applied to the prevention of opportunistic infections due to chemotherapy-induced immunosuppression in cancer patients.Citation44 A recent study showed that intratumoral Gammaproteobacteria in a mouse model of colon carcinoma conferred gemcitabine chemoresistance via metabolizing gemcitabine into inactive metabolite.Citation45 Intriguingly, the co-treatment with antibiotic ciprofloxacin dramatically reversed drug resistance in mice.Citation45 In consistent with this, several retrospective clinical studies also demonstrated that targeting cancer-promoting microbes with antibiotics improved gemcitabine response and prolonged survival time in patients with advanced pancreatic cancer.Citation46–48 Therefore, targeting the gut microbiota could be promising to optimize the outcomes of cancer treatment.Citation49–51 However, there are no available data by far about the effect of the gut microbiota modulation on ESCC therapy. In our current study, we found that F. nucleatum contributed to ESCC progression and chemoresistance via the enhanced secretion of chemotherapy-induced SASP, which further supports F. nucleatum as a potential therapeutic target in ESCC patients. It will be greatly desirable to explore the possible impact of antibiotic use on ESCC chemotherapy in the future studies.
In conclusion, our results indicated that chemotherapy-induced senescent ESCC cells was detrimental to the outcome of cancer therapy mainly due to the paracrine actions of the SASP. F. nucleatum promoted ESCC progression and chemoresistance via the enhanced secretion of chemotherapy-induced SASP. Our findings have for the first time established a crosstalk between F. nucleatum and chemotherapy-induced senescent cells and provided a novel insight into the mechanisms by which F. nucleatum promotes cancer development and chemoresistance. Therefore, targeting F. nucleatum may be a potential strategy for the treatment of ESCC in the future.
4. Materials and methods
4.1 Patients and sample collection
This study was approved by the Ethics Committee of Renmin Hospital of Wuhan University (Ethics No: WDRY2021-K062). Informed consent was obtained from all patients included in this study. To determine whether CDDP could induce cancer cell senescence in vivo, ESCC patients who had received CDDP-based neoadjuvant chemotherapy before surgical resection at Renmin Hospital of Wuhan University from January 2017 to December 2021 were collected. A total of 25 ESCC patients were included in this study. The pathological response to chemotherapy in these patients was evaluated by a certified pathologists according to the tumor regression grade system. Additionally, 20 ESCC patients who had not received chemotherapy prior to surgical resection were included as controls. Formalin-fixed paraffin-embedded tissues were obtained from the Department of Pathology of Renmin Hospital of Wuhan University. To perform 16S rRNA gene sequencing, 7 pairs of cancerous and adjacent normal tissues (at least 5 cm away from the tumor site) were obtained from ESCC patients who underwent surgical resection at Renmin Hospital of Wuhan University from January to April 2022. None of these patients received antibiotics treatment within the previous 3 months, and none were subject to other esophagus-related inflammatory diseases. All collected samples were fresh frozen and stored in liquid nitrogen. To clarify the correlation between F. nucleatum and the patient prognosis, ESCC patients who underwent surgical resection at Renmin Hospital of Wuhan University from January to December 2017 were collected. Patients who received antibiotics treatment within the previous 3 months were excluded. None of these patients were subject to other esophagus-related inflammatory diseases. A total of 107 patients with ESCC were included in this study. Tumor staging was performed according to the 8th edition of the Union for International Cancer Control TNM classification of malignant tumors. 107 pairs of formalin-fixed paraffin-embedded cancerous and adjacent normal tissues were obtained from the Department of Pathology of Renmin Hospital of Wuhan University. Prognostic information was collected by the medical record system and telephone follow-up.
4.2 Cell, bacterial strain, and co-culture system
Human ESCC cell lines KYSE-150 and ECA-109 obtained from Shanghai Cell Bank, Chinese Academy of Sciences were cultured in RPMI-1640 medium (Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Gaithersburg, MD, USA) and 1% penicillin/streptomycin (Beyotime, Shanghai, China) at 37°C in a humidified 5% CO2 atmosphere. The F. nucleatum strain ATCC 25,586 was purchased from the China General Microbiological Culture Collection Center (Beijing, China) and cultured in fastidious anaerobe broth (Tuopu biotechnology, Qingdao, China) at 37°C under anaerobic condition. For in vitro co-culture experiments, cells were washed with phosphate-buffered saline (PBS, Beyotime, Shanghai, China) and then incubated with F. nucleatum at a multiplicity of infection (MOI) of 100 in antibiotic-free RPMI-1640 medium supplemented with 10% FBS at 37°C under 5% CO2 for 24 h.
4.3 Reagents and antibodies
CDDP and KU55933 (a selective ATM kinase inhibitor) were purchased from MedChemExpress (MCE, Shanghai, China). CDDP was dissolved in sterile double distilled water at 1 mg/ml and stored at −80°C until dilution in the culture medium for treatment. KU55933 was dissolved at 10 mM in DMSO (Beyotime, Shanghai, China) and stored at −80°C. To inhibit cellular ATM activity in certain experiments, cells were pre-treated with KU55933 (10 μM) for 2 h. The primary antibodies including p53 (9282, CST, Danvers, MA, USA), p21 (2947, CST), γH2AX (9718, CST), cleaved Caspase-3 (9664, CST), cleaved PARP (5625, CST), Ki-67 (9027, CST), E-cadherin (14472, CST), N-cadherin (13116, CST), Vimentin (5741, CST), Snail (3879, CST), Slug (9585, CST), β-catenin (8480, CST), p65 (8242, CST), p-p65 (3033, CST), ATM (2873, CST), p-ATM (5883, CST), Chk2 (6334, CST), p-Chk2 (2197, CST), GAPDH (2118, CST), p16 (ab241543, Abcam, Cambridge, MA, USA), IL-6 (ab214429, Abcam), CXCL10 (ab214668, Abcam), MMP3 (ab52915, Abcam), IL-1α (16765–1-AP, Proteintech, Chicago, IL, USA), IL-1β (16806–1-AP, Proteintech), IL-8 (27095–1-AP, Proteintech), MyD88 (23230–1-AP, Proteintech), and TLR4 (19811–1-AP, Proteintech) were used to detect protein expression.
4.4 CCK-8 assay
Cell viability was detected using CCK-8 assay (Beyotime, Shanghai, China) based on the manufacturer’s instructions. Briefly, ESCC cells were seeded in 96-well plates at a density of 2.5 × 10Citation3 cells per well. At the indicated time points, 100 μl RPMI-1640 medium supplemented with 10 μl CCK-8 were added to each well and then incubated at 37°C for 1 h. The absorbance at 450 nm was measured by a microplate reader.
4.5 Colony formation assay
ESCC cells were seeded in 6-well plates at a density of 1 × 10Citation5 cells per well, cultured overnight, and treated with or without CDDP at the indicated concentrations for 96 h. Then, the cells were trypsinized and re-plated into 6-well plates at a density of 1 × 10Citation3 cells per well and cultured for 10 days. Colonies were fixed with 4% paraformaldehyde (Beyotime, Shanghai, China) for 15 min and stained with 0.1% crystal violet (Beyotime, Shanghai, China) for 15 min. The number of cell colonies was counted using Image J software (National Institutes of Health, Bethesda, MD, USA).
4.6 Cell migration and invasion assay
The transwell assay was conducted to detect the impact of the different CM on the migration and invasion of ESCC cells. For the migration assay, ESCC cells at a density of 5 × 10Citation4 cells per well were seeded in the upper compartment of the chamber (8 μm pore size, Corning, MA, USA) in serum-free RPMI-1640 medium. The lower compartment was filled with the different CM. After incubation at 37°C for 36 h, cells were fixed with 4% paraformaldehyde for 15 min and stained with 0.1% crystal violet for 15 min. The non-migrating cells on the upper surface of the chamber were completely removed with cotton swabs. The migrated cells on the lower surface were photographed under a light microscope (Olympus IX71, Tokyo, Japan) in five random fields at 200 × magnification, and the number of cells was directly counted. For the invasion assay, the upper compartment of the chamber was precoated with Matrigel (Corning, MA, USA). All other steps were the same as the migration assay.
4.7 EdU incorporation assay
The ability of cell proliferation was measured with EdU incorporation assay using BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 488 (Beyotime, Shanghai, China) according to the manufacturer’s instructions. In brief, at the indicated time points, cells were incubated with 10 μM EdU at 37°C for 2 h. Then, cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.3% Triton X-100 for 15 min. Subsequently, cells were incubated with Click Additive Solution for 30 min and stained with Hoechst 33342 for 10 min in the dark. Five random fields at 200 × magnification were captured under a fluorescent microscope (Olympus IX53, Tokyo, Japan).
4.8 Flow cytometry
The distribution of cell cycle was detected using the cell cycle detection kit (Multi Sciences, Hangzhou, China) in accordance with the manufacturer’s protocols. Briefly, cells were incubated with DNA staining solution and permeabilization solution at room temperature for 30 min in the dark. The DNA content was detected by flow cytometry using a FACS Calibur system (Becton Dickinson, Franklin Lakes, NJ, USA), and the data were analyzed using FlowJo FACS analysis software (TreeStar, Ashland, OR, USA). The FITC-Annexin V apoptosis detection kit I (BD Pharmingen, San Diego, CA, USA) was used to detect apoptosis based on the manufacturer’s protocols. Cells were suspended in FITC-Annexin V binding buffer and then stained with FITC-Annexin V and PI for 15 min at room temperature in the dark. The stained cells were analyzed using a flow cytometer (Beckman Coulter, Miami, FL, USA).
4.9 SA-β-gal staining
The activity of SA-β-Gal was determined using SA-β-Gal staining kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. Cells were fixed with the fixative solution and then stained with the staining solution overnight at 37°C in a non-CO2 incubator. Five random fields at 200 × magnification were captured under a light microscope (Olympus IX71, Tokyo, Japan).
4.10 RT-qPCR
Total RNA was extracted from cultured cells using Trizol reagent (Takara, Ohtsu, Japan) based on the manufacturer’s instructions. Complementary DNA was synthesized using the Reverse Transcription kit (Takara, Ohtsu, Japan). RT-qPCR was performed using the SYBR Green PCR Master Mix (Takara, Ohtsu, Japan) on a Bio-Rad CFX ConnectTM Real-Time PCR Detection System. GAPDH was used as an internal control. The relative mRNA expression of target gene was calculated by the 2−ΔΔCt method. All primer sequences were listed in .
4.11 Western blot
Total protein was isolated from harvested cells by RIPA lysis buffer (Beyotime, Shanghai, China). The protein extract concentration was determined using the BCA protein assay kit (Beyotime, Shanghai, China) in accordance with the manufacturer’s instructions. Equal amounts of protein were separated by 10% SDS-PAGE and then transferred to the polyvinylidene fluoride membrane (Millipore, MA, USA). After blocking with 5% nonfat milk at room temperature for 1 h, the membrane was incubated with the primary antibodies overnight at 4°C. The next day, the membrane was incubated with the suitable secondary antibodies for 1 h at room temperature. The protein signals were detected by enhanced chemiluminescence with ECL detection reagents on a Bio-Rad Detection System.
4.12 Immunofluorescent staining
Immunofluorescent staining was conducted as described previously.Citation52 Briefly, at the indicated time points, cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.2% Triton X-100 (Beyotime, Shanghai, China) for 15 min followed by blocking with 10% goat serum (Beyotime, Shanghai, China) at room temperature for 1 h. Then, cells were incubated with the primary antibodies overnight at 4°C and then incubated with Cy3-labeled secondary antibody (BOSTER, Wuhan, China) or Alexa Fluor 488-labeled secondary antibody (Beyotime, Shanghai, China) for 1 h at 37°C in the dark. Subsequently, cells were stained with 4,6-diamidino-2-phenylindole (DAPI, Beyotime, Shanghai, China) for 10 min. Five random fields at 400 × magnification were photographed under a fluorescent microscope (Olympus BX53, Tokyo, Japan).
4.13 Hematoxylin-eosin (H&E) and immunohistochemistry staining
Formalin-fixed paraffin-embedded tissues were performed for H&E and immunohistochemistry staining according to previously described protocols.Citation48 Briefly, the tissue sections were dewaxed, rehydrated, and boiled in citrate buffer for antigen retrieval followed by blocking with 10% goat serum. Then, the slides were incubated with the primary antibodies overnight at 4°C followed by incubation with the suitable secondary antibodies for 30 min at room temperature. The sections were observed using a light microscope (Olympus IX71, Tokyo, Japan) at 400 × magnification. All tissue sections were examined by a certified pathologist.
4.14 16S rRNA gene sequencing
For 16S rRNA gene sequencing, 7 pairs of fresh frozen cancerous and matched adjacent normal tissues of ESCC patients were processed by Shanghai Majorbio Bio-Pharm Technology Co., Ltd (Shanghai, China) where microbial genomic DNA was extracted, amplified, and sequenced following the standard procedures. Briefly, microbial genomic DNA was isolated using the E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, USA). The V3-V4 hypervariable regions of the bacterial 16S rRNA gene were amplified using universal primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806 R (5′-GGACTACHVGGGTWTCTAAT-3′). The amplicon sequencing was performed with the Illumina MiSeq platform (Illumina, San Diego, CA, USA). The raw sequencing data were first trimmed and quality-filtered to remove adaptor or low-quality sequences. Filtered reads were clustered into the operational taxonomic units (OTUs) assuming 97% similarity. According to the Silva 16S rRNA database, the taxonomic information of each representative sequence was annotated. MOTHUR software was used to evaluate the microbial α diversity indexes including the Simpson index, Shannon index, Chao 1 index, and Ace index. The differences in the α diversity between the two groups were analyzed by the Student’s t test. The β diversity analysis was used to compare the differences in species diversity among the groups via principal coordinate analysis (PCoA) and non-metric multidimensional scaling (NMDS) analysis based on weighted unifrac distance. The differences in the β diversity between the two groups were estimated by analysis of similarities (ANOSIM). The differential species between the two groups at the phylum and genus levels were compared using Wilcoxon rank-sum test.
4.15 FISH analysis
FISH was carried out to detect F. nucleatum in the 5-μm-thick sections of formalin-fixed and paraffin-embedded cancerous and paired adjacent normal tissues of ESCC patients as described previously.Citation53 A specific F. nucleatum-targeted 16S rRNA oligonucleotide probe (5′-CTT GTA GTT CCG C(C/T) TAC CTC-3′) labeled with Cy3 was synthesized by FOCOFISH (Guangzhou, China). Five random fields at 200 × magnification per sample were captured under a fluorescent microscope (Olympus BX53, Tokyo, Japan), and the number of F. nucleatum per field was counted by an observer blind to sample status.
4.16 F. nucleatum invasion assay
The invasion assay was performed as previously described.Citation22 In brief, ESCC cells were seeded in 6-well plates at a density of 1 × 10Citation5 cells per well, cultured overnight, and treated with or without CDDP at the indicated concentrations for 96 h. Before co-culture with F. nucleatum, cells were washed with PBS and cultured in antibiotic-free 1640-RPMI medium. Then, F. nucleatum (MOI = 100) was added to the cells followed by incubation at 37°C under 5% CO2 for 24 h. After incubation, the cells were washed with PBS and then incubated in serum-free fresh medium containing 200 μg/ml metronidazole and 300 μg/ml gentamicin for 1 h, which could completely eliminate extracellular F. nucleatum in the culture medium as verified by our in vitro experiment. After exposure to the antibiotics, the cells were washed with PBS and lysed with 250 μl of sterile distilled water at 37°C for 15 min. The lysates were plated on blood agar plates and cultured under anaerobic condition at 37°C for 72 h. The viable bacterial counts were quantified using Image J software (National Institutes of Health, Bethesda, MD, United States).
4.17 Collection of conditioned medium (CM)
ESCC cells were seeded in 6-well plates at a density of 1 × 10Citation5 cells per well, cultured overnight, and treated with or without CDDP at the indicated concentrations for 96 h. Then, cells were washed with PBS and cultured in serum-free 1640-RPMI medium. After 48 h, the CM was collected, centrifuged, and filtered to remove cell debris. The CM was concentrated 5-fold with a Centricon Centrifugal filter (3 kD, Millipore, Temecula, CA, USA). The concentration of the CM was normalized to the number of cells collected. The CM from CDDP-treated ESCC cells was defined as Sen CM, and the CM from untreated cells was defined as n-Sen CM. For in vitro co-culture experiments, ESCC cells treated with or without CDDP for 96 h were washed with PBS and co-cultured with or without F. nucleatum (MOI = 100) in antibiotic-free RPMI-1640 medium supplemented with 10% FBS at 37°C under 5% CO2 for 24 h. Then, cells were washed with PBS and cultured in serum-free 1640-RPMI medium for 48 h. The CM from F. nucleatum-infected or uninfected senescent ESCC cells (defined as Sen+F. nucleatum CM or Sen CM) and the CM from F. nucleatum-infected or uninfected non-senescent cells (as n-Sen+F. nucleatum CM or n-Sen CM) were harvested and then used for the following experiments.
4.18 Enzyme-linked immunosorbent assay (ELISA)
For cytokine analysis, the different CM was prepared as mentioned above. The concentrations of IL-1α, IL-1β, IL-6, IL-8, MMP3, and CXCL10 were measured respectively using IL-1α (KE00123, Proteintech, Chicago, IL, USA), IL-1β (KE00021, Proteintech), IL-6 (KE00139, Proteintech), IL-8 (KE00006, Proteintech), MMP3 (KE00160, Proteintech), and CXCL10 (ab83700, Abcam, Cambridge, MA, USA) ELISA kits following the manufacturer’s instructions. The absorbance at 450 nm was measured by a microplate reader. The protein levels were calculated according to the standard curve.
4.19 Neutral comet assay (single cell gel electrophoresis assay)
To evaluate the level of DNA damage in cells, neutral comet assay was carried out using the Comet AssayⓇ kit (R&D Systems) based on the manufacturer’s instructions. In brief, the cells were suspended at a density of 1 × 10Citation5 cells/ml in ice cold PBS, mixed with molten LMAgarose at a ratio of 1:10 at 37°C, and loaded to 2-well slides from the kit. The slides were immersed in pre-cooled lysis solution for 1 hour and soaked with neutral electrophoresis solution for 30 min at 4°C. Then, the slides were placed into horizontal electrophoresis tanks filled with fresh pre-cooled electrophoresis solution and electrophoresed at 21 volts for 45 min at 4°C in the dark. Subsequently, the slides were immersed in DNA precipitation solution and 70% ethanol for 30 min, respectively. The slides were stained with diluted SYBR Green I staining solution for 30 min in the dark and photographed under a fluorescent microscope (Olympus BX53, Tokyo, Japan). At least 50 cells per sample at 200 × magnification were randomly selected to score the olive tail moment using the Comet Score software.
4.20 Tumor xenograft experiments
All animal experiments were approved by the Laboratory Animal Welfare and Ethics Committee of Renmin Hospital of Wuhan University (IACCU Issue No: 20201203). Great efforts were made to minimize the suffering of the included animal. The effect of the CM from senescent cells on the proliferation of KYSE-150 cells was evaluated using the xenograft model. Two groups of mice were assessed for tumor development: 1) tumor-bearing mice treated with Sen CM and 2) tumor-bearing mice treated with n-Sen CM. Additionally, we also measured the impact of the CM from F. nucleatum-infected and uninfected senescent cells on KYSE-150 cell proliferation in vivo. Four groups of mice were assessed for tumor development: 1) tumor-bearing mice treated with Sen CM, 2) tumor-bearing mice treated with Sen+F. nucleatum CM, 3) tumor-bearing mice treated with n-Sen CM, and 4) tumor-bearing mice treated with n-Sen+F. nucleatum CM. An inoculum of 5 × 10Citation6 KYSE-150 cells were injected subcutaneously into the right flank of 5-week-old male BALB/c nude mice (5 per group). Next, the mice were intraperitoneally injected every other day with 5-fold concentrated CM from different treatment groups, four times in total. When the tumors reached a volume of 50 mm3, the mice were intratumorally injected every other day with CM from different treatment groups, four times in total. The tumor volume was calculated every 4 days using the following formula: Tumor volume (mmCitation3) = ½ × L × WCitation2, where L is the length and W is the width. The mice were euthanized on day 24, with tumors excised and weighed. The xenograft tissues were fixed with 4% paraformaldehyde and embedded in paraffin for H&E and immunohistochemistry staining.
4.21 Statistical analysis
All statistical analyses were performed using GraphPad Prism 8.0 except that the optimal cutoff value for F. nucleatum abundance was assessed using X-tile software. Categorical data were presented as numbers or proportions. Groups were compared using the chi-square test or Fisher’s exact test. Continuous variables were expressed as mean ± standard deviation. The two groups were compared using the Student’s t test, and multiple groups were compared using one-way analysis of variance (ANOVA) with Bonferroni correction. For time-to-event analyses, survival estimates were calculated using the Kaplan-Meier analysis, and the survival differences between the two groups were compared using the log-rank test. To obtain statistics, all studies were performed as three independent biological experiments unless otherwise stated. A statistically significant difference was defined as *p < 0.05, **p < 0.01, and ***p < 0.001.
Abbreviations
| ESCC | = | esophageal squamous cell carcinoma |
| CDDP | = | cisplatin |
| SASP | = | senescence-associated secretory phenotype |
| F. nucleatum | = | Fusobacterium nucleatum |
| SA-β-Gal | = | senescence-associated beta-galactosidase |
| CM | = | conditioned medium |
| FISH | = | fluorescence in situ hybridization |
| FadA | = | Fusobacterium adhesin A |
| MOI | = | multiplicity of infection |
Author contributions
Jian-Wei Zhang: Conceptualization, Methodology, Project administration, Funding acquisition, Writing – original draft. Dan Zhang: Data curation, Validation, Formal analysis, Visualization. Hai-Sen Yin: Investigation, Validation, Resources. Han Zhang: Investigation, Resources. Kun-Qiao Hong: Investigation, Software, Formal analysis. Jing-Ping Yuan: Resources, Data curation. Bao-Ping Yu: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Consent for publication
All authors have read and approved the final draft of the manuscript.
Ethics approval
This study was approved by the Ethics Committee of Renmin Hospital of Wuhan University (Ethics No: WDRY2021-K062). Informed consent was obtained from all patients included in this study. All animal experiments were approved by the Laboratory Animal Welfare and Ethics Committee of Renmin Hospital of Wuhan University (IACCU Issue No: 20201203).
Supplemental Material
Download Zip (6.9 MB)Acknowledgments
We appreciate the excellent technical assistance from pathologists at Renmin Hospital of Wuhan University.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
All sequencing data associated with this study have been uploaded to the NCBI (Sequence Read Archive) SRA database under the accession number PRJNA866879 (http://www.ncbi.nlm.nih.gov/bioproject/866879).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2023.2197836.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: gLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–25. doi:10.3322/caac.21660.
- Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Niksic M, Bonaventure A, Valkov M, Johnson CJ, Estève J, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosd with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. doi:10.1016/S0140-6736(17)33326-3.
- He S, Xu J, Liu X, Zhen Y. Advances and challenges in the treatment of esophageal cancer. Acta Pharm Sin B. 2021;11(11):3379–3392. doi:10.1016/j.apsb.2021.03.008.
- Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C, Campisi J, Collado M, Evangelou K, Ferbeyre G, et al. Cellular senescence: defining a path forward. Cell. 2019;179(4):813–827. doi:10.1016/j.cell.2019.10.005.
- Lee S, Schmitt CA. The dynamic nature of senescence in cancer. Nat Cell Biol. 2019;21(1):94–101. doi:10.1038/s41556-018-0249-2.
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46. doi:10.1158/2159-8290.CD-21-1059.
- Hernandez-Segura A, Nehme J, Demaria M. Hallmarks of cellular senescence. Trends Cell Biol. 2018;28(6):436–453. doi:10.1016/j.tcb.2018.02.001.
- Faget DV, Ren Q, Stewart SA. Unmasking senescence: context-dependent effects of SASP in cancer. Nat Rev Cancer. 2019;19(8):439–453. doi:10.1038/s41568-019-0156-2.
- Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5(1):99–118. doi:10.1146/annurev-pathol-121808-102144.
- Jackson JG, Pant V, Li Q, Chang LL, Quintas-Cardama A, Garza D, Tavana O, Yang P, Manshouri T, Li Y, et al. P53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer Cell. 2012;21(6):793–806. doi:10.1016/j.ccr.2012.04.027.
- Nacarelli T, Fukumoto T, Zundell JA, Fatkhutdinov N, Jean S, Cadungog MG, Borowsky ME, Zhang R. NAMPT inhibition suppresses cancer stem-like cells associated with therapy-induced senescence in ovarian cancer. Cancer Res. 2020;80(4):890–900. doi:10.1158/0008-5472.CAN-19-2830.
- Sun X, Shi B, Zheng H, Min L, Yang J, Li X, Liao X, Huang W, Zhang M, Xu S, et al. Senescence-associated secretory factors induced by cisplatin in melanoma cells promote non-senescent melanoma cell growth through activation of the ERK1/2-RSK1 pathway. Cell Death Dis. 2018;9(3):260. doi:10.1038/s41419-018-0303-9.
- Kim YH, Choi YW, Lee J, Soh EY, Kim JH, Park TJ. Senescent tumor cells lead the collective invasion in thyroid cancer. Nat Commun. 2017;8(1):15208. doi:10.1038/ncomms15208.
- Demaria M, O’leary MN, Chang J, Shao L, Liu S, Alimirah F, Koenig K, Le C, Mitin N, Deal AM, et al. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 2017;7(2):165–176. doi:10.1158/2159-8290.CD-16-0241.
- Ahmadinejad F, Bos T, Hu B, Britt E, Koblinski J, Souers AJ, Leverson JD, Faber AC, Gewirtz DA, Harada H. Senolytic-mediated elimination of head and neck tumor cells induced into senescence by cisplatin. Mol Pharmacol. 2022;101(3):168–180. doi:10.1124/molpharm.121.000354.
- Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science. 2021;371(6536):371. doi:10.1126/science.abc4552.
- Brennan CA, Garrett WS. Fusobacterium nucleatum — symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17(3):156–166. doi:10.1038/s41579-018-0129-6.
- Casasanta MA, Yoo CC, Udayasuryan B, Sanders BE, Umana A, Zhang Y, Peng H, Duncan AJ, Wang Y, Li L, et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal. 2020;13(641). doi:10.1126/scisignal.aba9157.
- Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, et al. Fusobacterium nucleatum Increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor−κB, and up-regulating expression of microRNA-21. Gastroenterology. 2017;152(4):851–66 e24. doi:10.1053/j.gastro.2016.11.018.
- Nomoto D, Baba Y, Liu Y, Tsutsuki H, Okadome K, Harada K, Ishimoto T, Iwatsuki M, Iwagami S, Miyamoto Y, et al. Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression via the NOD1/RIPK2/NF-κB pathway. Cancer Lett. 2022;530:59–67. doi:10.1016/j.canlet.2022.01.014.
- Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–63 e16. doi:10.1016/j.cell.2017.07.008.
- Liu Y, Baba Y, Ishimoto T, Tsutsuki H, Zhang T, Nomoto D, Okadome K, Yamamura K, Harada K, Eto K, et al. Fusobacterium nucleatum confers chemoresistance by modulating autophagy in oesophageal squamous cell carcinoma. Br J Cancer. 2021;124(5):963–974. doi:10.1038/s41416-020-01198-5.
- Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4(12):1798–1806. doi:10.1038/nprot.2009.191.
- Hwang HJ, Lee YR, Kang D, Lee HC, Seo HR, Ryu JK, Kim Y-N, Ko Y-G, Park HJ, Lee J-S. Endothelial cells under therapy-induced senescence secrete CXCL11, which increases aggressiveness of breast cancer cells. Cancer Lett. 2020;490:100–110. doi:10.1016/j.canlet.2020.06.019.
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. doi:10.1158/1078-0432.CCR-04-0713.
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-Cadherin/β-catenin signaling via its FadA Adhesin. Cell Host & Microbe. 2013;14(2):195–206. doi:10.1016/j.chom.2013.07.012.
- Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11(8):973–979. doi:10.1038/ncb1909.
- Acosta JC, Gil J. Senescence: a new weapon for cancer therapy. Trends Cell Biol. 2012;22(4):211–219. doi:10.1016/j.tcb.2011.11.006.
- Zhang JW, Zhang D, Yu BP. Senescent cells in cancer therapy: why and how to remove them. Cancer Lett. 2021;520:68–79. doi:10.1016/j.canlet.2021.07.002.
- Sieben CJ, Sturmlechner I, van de Sluis B, van Deursen JM. Two-step senescence-focused cancer therapies. Trends Cell Biol. 2018;28(9):723–737. doi:10.1016/j.tcb.2018.04.006.
- Sikora E, Bielak-Zmijewska A, Mosieniak G. Targeting normal and cancer senescent cells as a strategy of senotherapy. Ageing Res Rev. 2019;55:100941. doi:10.1016/j.arr.2019.100941.
- Prasanna PG, Citrin DE, Hildesheim J, Ahmed MM, Venkatachalam S, Riscuta G, Xi D, Zheng G, Deursen JV, Goronzy J, et al. Therapy-induced senescence: opportunities to improve anticancer therapy. J Natl Cancer Inst. 2021;113(10):1285–1298. doi:10.1093/jnci/djab064.
- Fletcher-Sananikone E, Kanji S, Tomimatsu N, Di Cristofaro LFM, Kollipara RK, Saha D, Floyd JR, Sung P, Hromas R, Burns TC, et al. Elimination of radiation-induced senescence in the brain tumor microenvironment attenuates glioblastoma recurrence. Cancer Res. 2021;81(23):5935–5947. doi:10.1158/0008-5472.CAN-21-0752.
- Hainaut P, Pfeifer GP. Somatic TP53 mutations in the era of genome sequencing. Cold Spring Harb Perspect Med. 2016;6(11):a026179. doi:10.1101/cshperspect.a026179.
- Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, Sawayama H, Kinoshita K, Ishimoto T, Iwatsuki M, et al. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res. 2016;22(22):5574–5581. doi:10.1158/1078-0432.CCR-16-1786.
- Yin H, Zhang J, Zhang H, Li Q, Qiu H, Hong K, Wang W, Xiao Y, Yu B. Fusobacterium nucleatum promotes proliferation in oesophageal squamous cell carcinoma via AHR/CYP1A1 signalling. FEBS J. 2022;290(3):837–854. doi:10.1111/febs.16619.
- Yamamura K, Izumi D, Kandimalla R, Sonohara F, Baba Y, Yoshida N, Kodera Y, Baba H, Goel A. Intratumoral Fusobacterium nucleatum levels predict therapeutic response to neoadjuvant chemotherapy in esophageal squamous cell carcinoma. Clin Cancer Res. 2019;25(20):6170–6179. doi:10.1158/1078-0432.CCR-19-0318.
- Liang M, Liu Y, Zhang Z, Yang H, Dai N, Zhang N, Sun W, Guo Y, Kong J, Wang X, et al. Fusobacterium nucleatum induces MDSCs enrichment via activation the NLRP3 inflammosome in ESCC cells, leading to cisplatin resistance. Ann Med. 2022;54(1):989–1003. doi:10.1080/07853890.2022.2061045.
- Birch J, Gil J. Senescence and the SASP: many therapeutic avenues. Genes Dev. 2020;34(23–24):1565–1576. doi:10.1101/gad.343129.120.
- Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313(5788):848–851. doi:10.1126/science.1127059.
- Zanzoni A, Spinelli L, Braham S, Brun C. Perturbed human sub-networks by Fusobacterium nucleatum candidate virulence proteins. Microbiome. 2017;5(1):89. doi:10.1186/s40168-017-0307-1.
- Sanders BE, Umana A, Lemkul JA, Slade DJ. Erratum for sanders et al., “FusoPortal: an interactive repository of hybrid MinION-Sequenced Fusobacterium genomes improves gene identification and characterization”. mSphere. 2018 ;3(4). doi:10.1128/mSphere.00379-18.
- Guo P, Tian Z, Kong X, Yang L, Shan X, Dong B, Ding X, Jing X, Jiang C, Jiang N, et al. FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J Exp Clin Cancer Res. 2020;39(1):202. doi:10.1186/s13046-020-01677-w.
- Teillant A, Gandra S, Barter D, Morgan DJ, Laxminarayan R. Potential burden of antibiotic resistance on surgery and cancer chemotherapy antibiotic prophylaxis in the USA: a literature review and modelling study. Lancet Infect Dis. 2015;15(12):1429–1437. doi:10.1016/S1473-3099(15)00270-4.
- Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357(6356):1156–1160. doi:10.1126/science.aah5043.
- Imai H, Saijo K, Komine K, Otsuki Y, Ohuchi K, Sato Y, Okita A, Takahashi M, Takahashi S, Shirota H, et al. Antibiotic therapy augments the efficacy of gemcitabine-containing regimens for advanced cancer: a retrospective study. cancer Manag Res. 2019;11:7953–7965. doi:10.2147/CMAR.S215697.
- Nakano S, Komatsu Y, Kawamoto Y, Saito R, Ito K, Nakatsumi H, Yuki S, Sakamoto N. Association between the use of antibiotics and efficacy of gemcitabine plus nab-paclitaxel in advanced pancreatic cancer. Medi (Baltimore). 2020;99(39):e22250. doi:10.1097/MD.0000000000022250.
- Mohindroo C, Hasanov M, Rogers JE, Dong W, Prakash LR, Baydogan S, Mizrahi JD, Overman MJ, Varadhachary GR, Wolff RA, et al. Antibiotic use influences outcomes in advanced pancreatic adenocarcinoma patients. Cancer Med. 2021;10(15):5041–5050. doi:10.1002/cam4.3870.
- Ting NL, Lau HC, Yu J. Cancer pharmacomicrobiomics: targeting microbiota to optimise cancer therapy outcomes. Gut. 2022;71(7):1412–1425. doi:10.1136/gutjnl-2021-326264.
- Liu L, Shah K. The potential of the gut microbiome to reshape the cancer therapy paradigm: a review. JAMA Oncol. 2022;8(7):1059. doi:10.1001/jamaoncol.2022.0494.
- Park EM, Chelvanambi M, Bhutiani N, Kroemer G, Zitvogel L, Wargo JA. Targeting the gut and tumor microbiota in cancer. Nat Med. 2022;28(4):690–703. doi:10.1038/s41591-022-01779-2.
- Paffenholz SV, Salvagno C, Ho YJ, Limjoco M, Baslan T, Tian S, Kulick A, de Stanchina E, Wilkinson JE, Barriga FM, et al. Senescence induction dictates response to chemo- and immunotherapy in preclinical models of ovarian cancer. Proc Natl Acad Sci U S A. 2022;119(5):119. doi:10.1073/pnas.2117754119.
- Kong C, Yan X, Zhu Y, Zhu H, Luo Y, Liu P, Ferrandon S, Kalady MF, Gao R, He J, et al. Fusobacterium nucleatum promotes the development of colorectal cancer by activating a Cytochrome P450/Epoxyoctadecenoic acid axis via TLR4/Keap1/NRF2 signaling. Cancer Res. 2021;81(17):4485–4498. doi:10.1158/0008-5472.CAN-21-0453.
