ABSTRACT
The anaerobic bacterium Fusobacterium nucleatum is significantly associated with human colorectal cancer (CRC) and is considered a significant contributor to the disease. The mechanisms underlying the promotion of intestinal tumor formation by F. nucleatum have only been partially uncovered. Here, we showed that F. nucleatum releases a metabolite into the microenvironment that strongly activates NF-κB in intestinal epithelial cells via the ALPK1/TIFA/TRAF6 pathway. Furthermore, we showed that the released molecule had the biological characteristics of ADP-heptose. We observed that F. nucleatum induction of this pathway increased the expression of the inflammatory cytokine IL-8 and two anti-apoptotic genes known to be implicated in CRC, BIRC3 and TNFAIP3. Finally, it promoted the survival of CRC cells and reduced 5-fluorouracil chemosensitivity in vitro. Taken together, our results emphasize the importance of the ALPK1/TIFA pathway in Fusobacterium induced-CRC pathogenesis, and identify the role of ADP-H in this process.
KEYWORDS:
Introduction
Colorectal cancer (CRC) is one of the most common cancers, ranked third in incidence (1.8 million new cases/year) and is the second leading cause of cancer-related mortality (881000 deaths).Citation1 Although the etiology of CRC remains unclear, intestinal inflammation and various genetic factors contribute to CRC development. However, the relatively low heritability of CRC emphasizes the importance of environmental factors in disease development. Such factors encompass lifestyle changes that have occurred in developed societies over the last century, potentially affecting intestinal microbiota composition (antibiotics, smoking, diet low in fibers, and consumption of highly processed foods).Citation2
Sequencing approaches have been applied to explore microbiome profiles and unveiling distinct taxonomic bacterial composition referred to as dysbiosis in patients with CRC when compared to healthy individuals.Citation3,Citation4 Furthermore, recent research has evidenced the role of several intestinal bacteria in the development and severity of numerous cancers and also in influencing the effectiveness of cancer therapies.Citation3,Citation4 The impact of Porphyromonas, Escherichia, Bacteroides, Streptococcus and Fusobacterium has been particularly investigated in CRC. These species are often enriched in tumor tissues compared to healthy adjacent mucosa.Citation5–8 Among these species, the gram-negative anaerobic bacterium Fusobacterium nucleatum stands out as the most prevalent bacterium associated with CRCCitation9,Citation10 across different stages and subgroups of the disease.Citation3,Citation4,Citation8,Citation9,Citation11–15 Numerous studies have consistently reported an enrichment F. nucleatum in CRC samples from populations of diverse geographical origins, including Europe, Asia and America highlighting a worldwide relevance.Citation3,Citation4,Citation8,Citation12,Citation15–17 Moreover, F. nucleatum presence in CRC patients is associated with poor patient prognosis and resistance to chemotherapy.Citation18,Citation19 In recent years, the impact of F. nucleatum on CRC has been extensively studied using cellular and animal models. Mechanistically, the F. nucleatum effector, Fusobacterium adhesin protein 2 (Fap2), has been reported to bind host epithelial cells by interacting with the tumor-specific sugar residue Gal-GalNAc, facilitating Fusobacterium localization and enrichment in CRC.Citation20 In addition, a study has demonstrated that the effector Fusobacterium adhesin A (FadA), expressed at the membrane of F. nucleatum, activates the Wnt/β-catenin signaling pathway leading to the induction of oncogenic and inflammatory responses.Citation21 FadA has also been reported to bind to E-cadherin and to induce DNA damage such as DNA Double-Strand breaks (DSBs), resulting in chromosomal instability and cancer development.Citation22 Finally, F. nucleatum modulates the tumor-immune microenvironment by inhibiting natural killer cell (NK) cytotoxicity through Fap2, which binds to the human immune inhibitory receptor T-cell immunoglobulin and ITIM domain (TIGIT).Citation23 Interestingly, it has been demonstrated that Fusobacterium promotes the proliferation and reduces the chemosensitivity of CRC cells. This occurs through the activation of the transcription factor NF-κB, by Fusobacterium lipopolysaccharide (LPS), leading to the dysregulation of the autophagy and the anti-apoptotic pathways.Citation18,Citation19,Citation24 NF-κB contributes to intestinal homeostasis and is induced by microbial ligands recognized by Pattern recognition receptors (PRRs). Indeed, dysregulation of the PRR-dependent NF-κB signaling pathway has been linked to chronic inflammation, as well as dysfunction in barrier and tissue repair mechanisms. These dysfunctions often lead to excessive repair responses and cell proliferation, which are involved in various stages of carcinogenicity.Citation25–27 Despite being one of the most extensively studied CRC-related bacteria, the mechanisms through which F. nucleatum activates NF-κB and consequently drives CRC pathogenesis have not been fully characterized.
Recently, Alpha kinase 1 (ALPK1), a new PRR that senses ADP-heptose (ADP-H) produced by pathogenic and commensal gram-negative bacteria, has been described.Citation28–31 The binding of ADP-H to ALPK1 induces TIFA phosphorylation and subsequent NF-κB activation, which has been suggested to play a role in the pro-inflammatory response against pathogens.Citation29,Citation32,Citation33 A recent study showed that ALPK1 induces ICAM-1 expression upon F. nucleatum exposure, increasing CRC cell adhesion to endothelial cells.Citation34 Interestingly, ALPK1 regulates intestinal homeostasis in a mouse model of colitisCitation35 and single-nucleotide polymorphisms (SNPs) in this gene are associated with a range of chronic inflammatory diseases and various cancers, including CRC.Citation36–40 A study has shown that TIFA is involved in the Aurora A- and NF-κB-dependent expression of anti-apoptotic factors involved in leukemic cell growth and chemoresistance in acute myeloid leukemia.Citation41 More importantly, the activation of the ALPK1-TIFA-NF-κB axis is directly linked to pro-oncogenic processes, promoting replication stress and DNA damage in gastric cells.Citation42 Collectively, these studies suggest that ALPK1 play a role in promoting carcinogenesis; however, the specific pro-oncogenic processes involved and their connection with gut bacteria have not been definitively established.
In the present study, we showed that F. nucleatum releases molecules into its microenvironment, which activate NF-κB in intestinal epithelial cells (IECs) independently of TLRs and Nucleotide-binding oligomerization domain-containing protein (NOD) receptors, and through the ALPK1/TIFA pathway. This ALPK1/TIFA-dependent activation of the NF-κB phenotype was conserved among all fusobacterial species tested and acted synergistically with the butyrate produced by these bacteria. We investigated the impact of ALPK1/TIFA activation by F. nucleatum supernatant on HT-29 CRC cells and observed that induction of this pathway increased the expression of two anti-apoptotic genes and reduced the chemosensitivity of CRC cell to 5-fluorouracil. Taken together, our study provides novel evidence suggesting that the ALPK1/TIFA pathway is associated with the remote effect of Fusobacterium on pro-oncogenic processes.
Results
Fusobacterium nucleatum releases molecules that induce NF-κB activity in IECs through ALPK1/TIFA/TRAF6 pathway, independently of TLRs and NODs
Previous studies have established that F. nucleatum is a potent activator of the pro-inflammatory transcription factor NF-κB in a wide range of cell-lines.Citation34,Citation43 Exposure to F. nucleatum leads to the activation of TLRs, NODs, and ALPK1, resulting in NF-κB responses in various cell types; however, the impact of the molecules released in the microenvironment by the bacterium has not been characterized.Citation34,Citation44–46 We used culture supernatants from F. nucleatum subsp. nucleatum (DMS15643) and found that this bacterium released molecules capable of activating NF-κB in a reporter system expressed in the intestinal epithelial cell (IEC) line HT-29 (HT-29-NF-κB cells) (). Additionally, F. nucleatum supernatant activated NF-κB in HEK cells bearing the same reporter system (HEK-NF-κB). Importantly, this HEK cell line lacks expression of TLR2 and TLR4, which are linked to F. nucleatum’s major activity in CRC ().Citation18,Citation19,Citation24,Citation34,Citation44,Citation45 To investigate whether the remaining TLRs expressed in HEK-NF-κB were responsible for the NF-κB response to F. nucleatum supernatants, we deleted the gene encoding the adaptor protein MYD88 (MYD88-/-), a key protein in TLR signaling pathways. Interestingly, when incubated with F. nucleatum supernatants, both HEK-NF-κB WT and MYD88-/- cells exhibited similar NF-κB activity, indicating that the supernatant induced NF-κB activation via a MYD88-independent pathway (). Altogether, our results strongly demonstrate that F. nucleatum molecules released into the microenvironment activate NF-κB independently of MYD88 and TLRs.
Figure 1. F. nucleatum activates NF-κB in HT-29 independently of MyD88 and NOD1. (a) Average NF-κB activity in HT-29-NFκB reporter system (10% vol/vol), induced by F. nucleatum supernatant or control media for 24 h. (b) HEK-NFκB reporter (WT, black bars) and deleted for MYD88 (MYD88−/−, gray bars) cells were incubated with F. nucleatum supernatant or control media for 24 h. (c) Treatment of HEK-NFκB reporter cells deleted for MYD88−/− with control media, F. nucleatum supernatant or the NOD1 ligand (IE-DAP) for 24 h in presence or absence of a NOD1 inhibitor (ML130). NF-κB activation was measured by SEAP secretion and expressed as mean ± SD fold change toward unstimulated cells. Data represent ≥ 3 independent experiments performed in technical duplicate or triplicate. Data analysis: Mann-Whitney test was used, ****P < .0001; ***P < .001; **P < .01; *P < ,05; P < .05 was considered as not significant (ns).
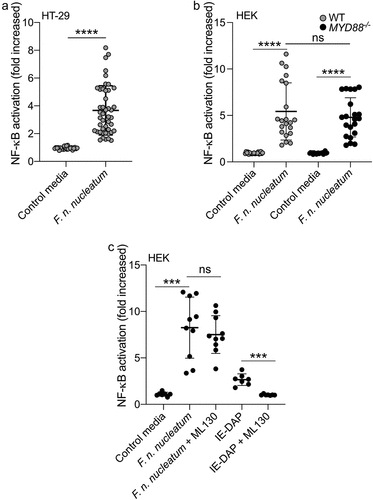
NOD1 and NOD2-dependent signaling pathways are reported to induce NF-κB activation in response to fusobacterial infections.Citation47 We excluded the involvement of NOD2 in F. nucleatum-dependent NF-κB activation, as this receptor is not expressed in HEK cells.Citation48 To investigate the role of NOD1, we treated MYD88−/− HEK cells with a specific NOD1-inhibitor (ML130). As expected, ML130 treatment completely abolished the activation of NF-κB by the purified NOD1 ligand IE-DAP. However, it did not affect NF-κB activation by F. nucleatum supernatant, indicating that NOD1 was not involved in the F. nucleatum-dependent activation of NF-κB ().
ALPK1 serves as an important cytosolic receptor in the innate immune response to gram-negative bacteria. Previous studies have demonstrated its activation upon exposure to F. nucleatum, Citation34 which lead us to hypothesize that ALPK1 might be involved in the NF-κB activation induced by F. nucleatum supernatant. ALPK1 senses intracellular ADP-H, leading to TIFA phosphorylation and consequently TIFA and TRAF6 oligomerization, resulting in the downstream activation of NF-κB.Citation28,Citation30 To validate our hypothesis, we assessed NF-κB activation by F. nucleatum supernatant in WT, ALPK1−/−, TIFA−/− and TRAF6−/− HEK cells. Our results demonstrated that the NF-κB response was dependent on the presence of ALPK1, TIFA, and TRAF6 (). Furthermore, we confirmed the critical role of this pathway by ectopically expressing TIFA in HEK TIFA−/− cells, which successfully restored NF-κB activation by F. nucleatum supernatant ().
Figure 2. F. nucleatum supernatant activates NF-κB via ALPK1, TIFA and TRAF6. (a) WT (gray dots), ALPK1−/− (green dots), TIFA−/− (red dots) and TRAF6−/− (bleu dots) HEK NF-κB-reporter cells were stimulated with F. nucleatum supernatant or control media for 24 h. NF-κB activation was measured by SEAP secretion and expressed as mean ± SD fold change toward unstimulated cells. (b) TIFA−/− HEK NF-κB-reporter cells transfected (gray dots) or not (red dots) with pTIFA-FLAG and stimulated with F. nucleatum supernatant or control media for 24 h. NF-κB activation was measured by SEAP secretion and expressed as mean ± SD fold change toward unstimulated cells. (c,d) TIFA-GFP HeLa cells were treated with control siRNA or ALPK1 specific siRNA prior to transfection with empty pCMV or pCMV-ALPK1 and were left unstimulated (control) or stimulated with ADP-H (10−6 M), F. nucleatum supernatant or control media for 24 h. Representative pictures of cells with TIFAsomes after 30 min of stimulation are in C (scale bar: 20 μm) and the graph showing the TIFAsomes quantification per cell in each condition is in d (control siRNA in black bars, siRNA ALPK1 in white bars and siRNA ALPK1 + pALPK1 in gray bars). (e) WT (gray dots) and TIFA−/− (red dots) HT-29 NF-κB reporter cells were stimulated with F. nucleatum supernatant or control media for 24 h. NF-κB activation was measured by SEAP secretion and expressed as mean ± SD fold change toward unstimulated cells. (f) CXCL8 relative expression to GAPDH in WT (black bars) and TIFA−/− (red bars) in HT29 stimulated with stimulated with ADP-H (10−6M), F. nucleatum supernatant or control media for 6 h expressed as 2−ΔΔCt toward unstimulated cells. g. IL-8 ELISA performed on WT (gray dots) and TIFA−/− (red dots) cells unstimulated (control) or treated with F. nucleatum supernatant or control media for 24 h. IL8 concentration was expressed in pg/ml. Data represent ≥ 3 independent experiments performed in duplicate or triplicate. Statistical significance was assessed using Mann-Whitney test (B, E-G) or one-way ANOVA followed by Tukey’s multiple comparisons test (A and D, for D, the samples were compared to their respective control transfected with control siRNA, siRNA ALPK1, siRNA ALPK1 + ALPK1). ****P < .0001; ***P < .001; **P < .01; *P < .05; P < .05 was considered as not significant (ns).
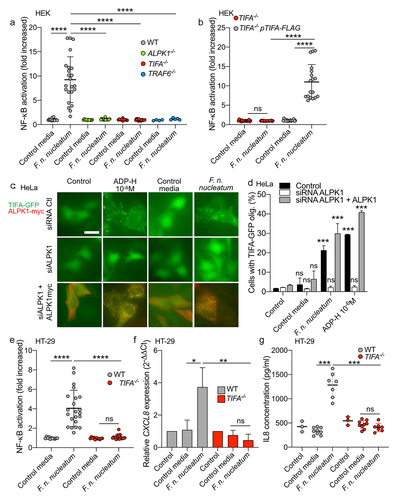
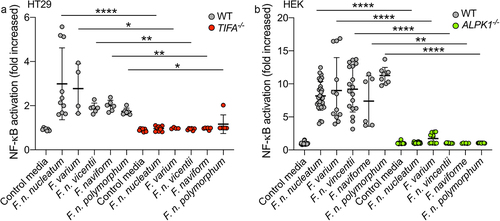
Upon stimulation of ALPK1 by its ligand, TIFA is phosphorylated, leading to the oligomerization of TIFA proteins into large structures called TIFAsomesCitation28,Citation30. Therefore, we assessed the impact of F. nucleatum supernatant on TIFAsome formation and investigated the role of ALPK1 in this process. To quantify TIFAsome formation, we used cells stably expressing GFP-tagged TIFA (TIFA-GFP) and transfected them with control or ALPK1-targeting siRNAs for gene silencing. In this experimental setup, we observed that both the bacterial supernatant and synthetic ADP-H induced TIFAsome formation, and that this phenotype was abolished in ALPK1-depleted cells. Furthermore, TIFAsome formation was restored in ALPK1-depleted cells upon rescue with an ectopically transfection of an ALPK1 cDNA construct (). To establish the relevance of these findings in intestinal epithelial cells, we showed that knocking out TIFA in HT-29 cells resulted in the abrogation of NF-κB activation by F. nucleatum supernatant and the downstream induction of IL-8 (encoded by CXCL8) at both the transcriptional and protein levels (). Moreover, ADP-H and F. nucleatum activated both NF-κB response and CXCL8 expression in the HCT116 cell-line, suggesting that the ALPK1 pathway is functional in other IECs (Supplementary Figure S1). Interestingly, we observed that the activation of the ALPK1/TIFA axis was not specific to F. nucleatum subsp. nucleatum, as all the fusobacterial species and strains tested exhibited similar ALPK1 and TIFA-dependent activation of NF-κB (). Taken together, our results provide strong support for the role of the ALPK1/TIFA/TRAF6 pathway in the activation of NF-κB by Fusobacterium culture supernatants in the intestinal context.
Figure 3. Fusobacterium species activate NF-κB via the ALPK1-TIFA pathway. (a) WT (gray dots) and TIFA−/− (red dots) HT-29 NF-κB-reporter cells were stimulated with supernatants derived from different Fusobacterium spp. Or control media for 24 h. (b) WT (gray dots) and ALPK1−/− (green dots) HEK NF-κB-reporter cells were incubated with supernatants derived from different Fusobacterium spp. Or control media for 24 h. NF-κB activation was measured by SEAP secretion and expressed as mean ± SD fold change toward unstimulated cells. Data represent ≥ 3 independent experiments performed in triplicate. Data analysis: Mann-Whitney test was used, ****P < ,0001; ***P < ,001; **P < ,01; *P < ,05; P < 0.05 was considered as not significant (ns).
Fusobacterium nucleatum releases in its microenvironment a NF-κB-activating molecule that has the biological features of the ALPK1 ligand, ADP-heptose
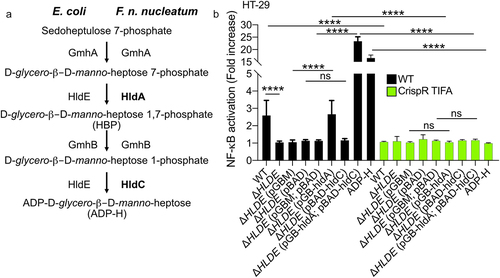
ADP-H and heptose-1,7-bisphosphate (HBP) are metabolites produced by gram-negative bacteria that trigger the ALPK1/TIFA pathway and consequently activate NF-κB.Citation28,Citation29 These molecules are intermediates in the heptose biosynthesis pathway, involved in LPS synthesis.Citation29 The KEGG database and genomic analysis revealed the presence of genes encoding enzymes (GmhA, HldA, HldC, and GmhB) involved in the LPS pathway in the genome of F. nucleatum ( and Supplementary Figure S2). Among these, HldA functions as a heptokinase, while HldC acts as an ADP-transferase enzyme, producing HBP and ADP-H respectively. In E. coli, both these enzymatic activities are carried out by the bifunctional enzyme HldE.Citation29 We took advantage of an hldE deletion mutant of E. coli (ΔhldE), in which we expressed F. nucleatum hldA and hldC to determine whether these enzymes were functional. Since ADP-H is not secreted by E. coli, bacterial lysates were used. Our results showed that lysates from WT and ΔhldE E. coli expressing hldA or both hldA and hldC from F. nucleatum potently activated NF-κB in HT-29 cells (), whereas the lysate of ΔhldE mutant did not, confirming the functionality of both enzymes. The mechanism was mediated by the ALPK1-TIFA pathway, as shown by the impaired NF-κB activation observed in HT-29 TIFA−/− cells treated with bacterial lysates from WT, ΔhldE alone, ΔhldE phldA-, and ΔhldE phldA/phldC-expressing E. coli (). Notably, the expression of both enzymes was required for optimal NF-κB activation, indicating that ADP-H is a more potent activator than HBP. Altogether, these results show that the expression of F. nucleatum hldA or both hldA/hldC in E. coli is sufficient to activate NF-κB in a TIFA-dependent manner ().
Figure 4. F. nucleatum enzymes HldA (Fn1786) and HldC (Fn0930) from the heptose biosynthesis pathway are functional. (a) Schematic view of the heptose biosynthesis pathway. (b) HT-29-NF-κB reporter WT (black bars) or TIFA−/− (green bars) cells were stimulated for 24 h with lysates from E. coli; ΔhldE; ΔhldE transformed with plasmid controls (pBAD and/or pGB), with hldA (pGB-hldA), with hldC (pBAD-hldC), with hldA (pGB-hldA) and hldC (pBAD-hldC) from F. nucleatum or ADP-H (10−6 M). NF-κB activation was measured by SEAP secretion and expressed as mean (%) ± SD fold change toward supernatant-stimulated cells. Data represent ≥ 3 independent experiments performed in triplicate. Data analysis: one-way ANOVA followed by Tukey’s multiple comparisons test was used, ****P < .0001; ***P < .001; **P < .01; *P < .05; P <.05 was considered as not significant (ns).
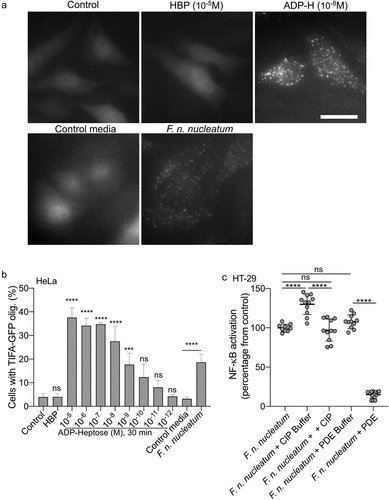
In contrast to ADP-H, HBP can indirectly and weakly activate the ALPK1/TIFA pathway after undergoing intracellular processing by host adenyltransferases into ADP-H 7-P.Citation29 To assess the contribution of HBP or ADP-H in the F. nucleatum activation of the ALPK1 pathway, we exploited the fact that, unlike ADP-H, HBP is unable to induce the formation of TIFAsomes within a 30-minutes of treatment.Citation28 Within this stimulation period, we observed that supernatants from F. nucleatum and synthetic ADP-H induced the formation of TIFAsomes, while HBP failed to do so (). The kinetics of TIFAsome formation induced by F. nucleatum were consistent with those of ADP-H, but not HBP. ADP-H exhibits partial resistance to calf intestinal alkaline phosphatase (CIP) but is sensitive to phosphodiesterase (PDE) from C. adamanteus, whereas HBP is only sensitive to CIP treatment.Citation49 To further confirm the role of ADP-H in F. nucleatum-induced NF-κB activation and exclude the contribution of HBP, we analyzed the effects of CIP and PDE treatments on bacterial culture supernatants. We showed that the NF-κB-activating molecule secreted by F. nucleatum was partially resistant to CIP and entirely sensitive to PDE (). Altogether, our findings support the conclusion that ADP-H, and not HBP, is responsible for the ALPK1-dependent activation of NF-κB byF. nucleatum supernatant.
Figure 5. F. nucleatum NF-κB activating molecule has the biological features of the ALPK1 ligand ADP-heptose. (a,b) cells were treated with digitonin and non-inoculated control medium, F. nucleatum supernatants, HBP (10−5M) or ADP-H (10−8M). in A: Representative pictures of cells with TIFAsomes at 30 min in TIFA-GFP-expressing HeLa cells (scale bar: 20 μm). In B, quantification of TIFAsomes in each condition as in a from three independent experiments performed in triplicate. (c) F. nucleatum supernatant was untreated, treated with calf intestine alkaline phosphatase (CIP), with C. adamanteus phosphodiesterase (PDE) or their respective buffer (CIP buffer and PDE buffer) prior to stimulation of HT-29-NF-κB reporter cells for 24 h. NF-κB activation was measured by SEAP secretion and results were expressed as mean change (%) ± SD toward F. nucleatum supernatant-stimulated cells. Data represent ≥ 3 independent experiments performed in triplicate. Statistical significance was assessed using one-way ANOVA followed by Tukey’s multiple comparisons test (for B, the samples were compared to their respective control: control for ADP-H and HBP and control media for F. nucleatum supernatants). ****P < ,0001; ***P < ,001; **P < ,01; *P < ,05; P < 0.05 was considered as not significant (ns).
Release of ADP-heptose and butyrate by fusobacterium synergize the ALPK1-dependentactivation of NF-κB by fusobacterium
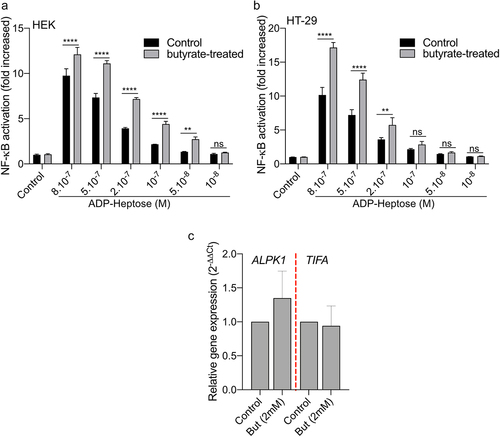
Fusobacterium species produce short-chain fatty acids, including butyrate, which can have a significant impact on host responsesCitation50–52(Supplementary Table S1). Butyrate has been reported as a synergistic inducer of NF-κB responses when combined with stimulation of TLRs and NODs.Citation53–55 In light of this, we investigated the potential synergy in NF-κB activation between butyrate and ALKP1-dependent signaling. We treated both the HT-29- and HEK-NF-κB reporter systems with butyrate (2 mM) and various concentrations of ADP-H (ranging from 8.10−7 to 10−8M) (). Butyrate alone did not induce NF-κB activation in either of the cell line. However, addition of butyrate (2 mM) to different concentrations of ADP-H led to a significant increase of NF-κB activation, nearly doubling the response compared to ADP-H stimulation alone (). It is well established that butyrate has a broad impact on the expression of host genes. We thus analyzed the expression of ALPK1 and TIFA following butyrate stimulation and found that butyrate had no effect on the transcription of these two genes (). Our results thus indicate that butyrate synergize with ADP-H to activate the NF-κB response.
Figure 6. Butyrate synergizes with ADP-H-dependent activation of NF-κB.A-B HEK (a) and HT-29 (b) NF-B-reporter cells were left untreated (control) or stimulated with ADP-H (10−8M to 8.10−7M) with (gray bars) or without (black bars) butyrate (2 mM). NF-κB activation was measured by SEAP secretion and expressed as the mean ± SD fold change toward unstimulated cells. Data represent three independent experiments performed in triplicate. (c) Real-time qPCR (RT-qPCR) showing ALPK1 and TIFA gene expression in HT-29 cells stimulated with 2 mM butyrate for 6 h. Results were normalized to GAPDH and expressed as 2−ΔΔCt toward unstimulated cells. Data represent three independent experiments performed in triplicate. Statistical significance was assessed using two-way ANOVA followed by Tukey’s multiple comparisons test. ****P < ,0001; ***P < ,001; **P < ,01; *P < ,05; P < 0.05, was considered as not significant (ns).
F. nucleatum supernatant promotes CRC cells proliferation and the expression of anti-apoptotic but not autophagic pro-cancerous genes via the ALPK1/TIFA pathway.
Recent studies have shown that Fusobacterium promotes CRC development and chemoresistance in a cell contact-dependent manner by targeting the innate immune receptor pathway TLR4/NF-κB. This process leads to the upregulation of miR21-dependent IEC proliferation, increased expression of the anti-apoptotic gene BIRC3, and activation of the autophagy pathway.Citation18,Citation19,Citation24 Here, we aimed to investigate whether secreted ADP-H might also contribute to the oncogenic effects of F. nucleatum, without need for direct cell contact, through the ALPK1/TIFA-mediated activation of NF-κB. To explore this hypothesis, we assessed whether F. nucleatum supernatant could mitigate the cytotoxicity induced by the anti-carcinogenic drug 5-Fluoroucil (5-Fluo) in WT and TIFA−/− HT-29 cells (). Our results showed that F. nucleatum supernatant reduced the cytotoxicity induced by 5-Fluo in HT-29 WT cells, albeit with a diminished effect in HT-29 TIFA−/− cells. Notably, treatment with the ALPK1 ligand, ADP-H, alone did not recapitulate the effect observed with the bacterial supernatant. Furthermore, F. nucleatum supernatant promoted the proliferation of HT-29 WT cells but had no impact on TIFA−/− cell (). Our data suggest that F. nucleatum supernatant contributes to the proliferation and chemoresistance of CRC cells in vitro, partially through the ALPK1-TIFA pathway.
Figure 7. F. nucleatum increases HT-29 proliferation and activates anti-apoptotic but not autophagic genes expression via the TIFA pathway. (a) WT (left) or TIFA−/− (right) HT-29 cells were left untreated (control), stimulated with ADP-H (10−6M), control media or F. nucleatum supernatant prior treatment with 5-fluoroucil (5-Fluo, 100 μM, gray bars) or without treatment (black bars). Cell viability HT-29 cell viability was monitored with MTS assay and normalized to the MTS response of untreated cells. B-E. Real-time qPCR (RT-qPCR) showing BIRC3 (b), TNAIP3 (c), ICAM-1 (d), ATG7 (e), ULK1 (f) and CCND1 (g) relative expression to GAPDH in WT (left) or TIFA−/− (right) HT-29 cells, untreated (control), stimulated with ADP-H (10−6M), control media or F. nucleatum supernatant for 6 h. Results were normalized on GAPDH and expressed as 2−ΔΔCt toward unstimulated cells. Data represent ≥ 3 independent experiments. Statistical significance was assessed using one-way ANOVA followed by Tukey’s multiple comparisons test. ****P < .0001; ***P < .001; **P < .01; *P < .05; P <.05 was considered as not significant (ns).
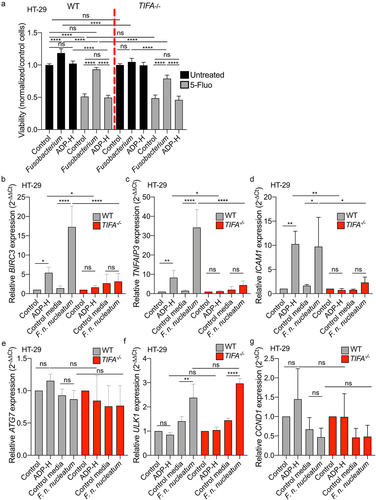
F. nucleatum has been reported to promote CRC carcinogenesis by influencing various pathways, including autophagy signaling, anti-apoptotic responses, inflammation, cell growth and CRC cell adhesion.Citation18,Citation19,Citation21,Citation34,Citation45 To gain further insights into the mechanisms involved in F. nucleatum-mediated CRC pathogenesis, we investigated the role of the ALPK1-TIFA axis in these targeted pathways. We thus assessed the impact of F. nucleatum supernatant on the expression of selected genes involved in autophagy (ULK1 and ATG7), cell growth (CCDN1), cell adhesion (ICAM1) and anti-apoptotic responses (BIRC3 and TNFAIP3). WT and TIFA−/− HT-29 cells were incubated for 6 h with F. nucleatum supernatants, non-inoculated control medium, or ADP-H prior to RNA extraction and RT-qPCR (). Our data revealed that ATG7 and CCND1 expression remained unaffected by F. nucleatum supernatant in HT-29 cells, regardless of the genetic background. F. nucleatum supernatant increased ULK1 expression in both WT and TIFA−/− cells, while ADP-H failed to do so. These results confirm the published resultsCitation18 and suggest that F. nucleatum activates the autophagic pathway independently of ALPK1/TIFA. In contrast, our data showed a significant TIFA-dependent upregulation of BIRC3, TNFAIP3 and ICAM1 expression induced by both F. nucleatum supernatant and ADP-H. Importantly, this enhanced transcription was TIFA-dependent (). These findings confirm the induction of ICAM1 expression by F. nucleatum and highlight that it is mediated by ADP-H released by the bacterium.Citation34 Altogether, our results demonstrate that F. nucleatum promotes the expression of anti-apoptotic and adhesion genes involved in Fusobacterium-mediated oncogenic mechanisms.Citation19,Citation34 This promotion occurs in a TIFA-dependent manner, underscoring the relevance of this pathway in CRC pathogenesis.
Discussion
Colorectal cancer (CRC) is a multifactorial disease influenced by both genetic and environmental factors.Citation56 Recent discoveries have placed the gut microbiota at the forefront of cancer research.Citation56,Citation57 Notably, metagenomic studies comparing the microbiomes of healthy individuals with those of CRC patients have unveiled correlations between microbiota composition and CRC outcome.Citation3,Citation8,Citation11–13,Citation16,Citation17 These studies have revealed significant microbiota signatures, with over 20 bacterial species, primarily originating from the oral cavity ecosystem, associated with various aspect of CRC including its development, severity, and resistance to chemotherapy.
Fusobacterium is implicated in the promotion of oncogenesis through a multitude of mechanisms, including DNA damage, hyperproliferation, malignant transformation in epithelial cells, and the development of chemoresistance.Citation18,Citation21,Citation22,Citation24 Recent studies have highlighted the role of Fusobacterium in driving CRC development and enhancing chemoresistance. These effects involved the activation of the innate immune receptor pathway TLR4/NF-κB, resulting in the upregulation of miR21-dependent IEC proliferation and the expression of the anti-apoptotic gene BIRC3 along with the activation of the autophagy pathway.Citation18,Citation19,Citation24 These studies underscore the role of TLR4-dependent signaling pathways in various stages of carcinogenesis. However, apart from TLR4, only a limited number of studies have explored the connection between PRR and their role in carcinogenesis concerning intestinal bacteria, such as F. nucleatum. Citation25–27
In our study, we report that the CRC-associated bacterium F. nucleatum releases extracellular heptose-related metabolites capable of activating NF-κB in vitro through the ALPK1/TIFA signaling pathway. Importantly, this phenotype is not unique to F. nucleatum alone, as it is conserved among other members of the Fusobacterium genus associated with CRC, such as F. varium and F. naviforme ().Citation58 Our work, along with previous studies, has unveiled that both commensal and pathogenic bacteria, including Akkermansia, Neisseria and Campylobacter share the capacity to release extracellularly heptose-related molecules.Citation31,Citation33,Citation59 Of note, this feature was not shared by a wide range of supernatants derived from intestinal Gram negative bacteria.Citation31 Through our results, we identified butyrate, which is abundantly produced by fusobacterial species, as a synergistic molecule that enhances the NF-κB response to heptose. Our data strongly suggest that the molecule responsible for ALPK1 activation is related to heptose, specifically ADP-H. This conclusion is supported by our demonstration that the expression of F. nucleatum enzymes involved in LPS biogenesis, hldA (responsible for HBP production) or both hldA/hldC (for HBP production followed by its modification to ADP-H) in ΔHld E. coli, is sufficient to activate NF-κB in a TIFA-dependent manner. Notably, the expression of both hldA and hldC is necessary for optimal NF-κB activation, underscoring the potency of ADP-H as the primary activator. Along the same line, ADP-H initiates rapid TIFAsome formation, whereas HBP necessitates intracellular processing by host adenyltransferases into ADP-H-7-P to achieve the same effect.Citation28,Citation29 Our assessment of TIFAsome formation in permeabilized cells 30 min after incubation of with F. nucleatum supernatant is a characteristic specific to the ALPK1-ligand ADP-H.Citation28 Moreover, our experiments have confirmed that the F. nucleatum NF-κB-activating compound, like ADP-H, undergoes hydrolysis by phosphodiesterase, providing strong evidence that the released compound is indeed ADP-H.Citation49
The central transcription factor NF-κB plays a pivotal role in inflammation and is associated with tumor promotion in colon cancers.Citation60 Prior research has unveiled the multifaceted involvement of F. nucleatum in CRC development and chemoresistance, with many of these mechanisms implicating NF-κB activation. F. nucleatum induces the expression of IL8, a pro-inflammatory and pro-angiogenic chemokine. IL8 activation triggers the PI3K and MAPK signaling pathways, fostering an oncogenic environment that promotes cell proliferation and survival.Citation61,Citation62 Upon F. nucleatum infection, ALPK1 activation leads to the expression of ICAM-1, which facilitates CRC cell adhesion to endothelial cells, promoting metastasis.Citation34,Citation63 F. nucleatum-induced NF-κB activation leads to the upregulation of BIRC3, an anti-apoptotic gene involved in IEC proliferation, and its dysregulation is associated with tumor cell growth and chemoresistance.Citation19,Citation64,Citation65 Through TLR4, F. nucleatum triggers the activation of ULK1 and ATG7, both playing crucial roles during the initial stages of autophagy. Autophagy is a cellular pathway important for maintaining intestinal homeostasis, promoting cell survivalCitation66 and potentially supporting cancer development and cell chemoresistance. By activating PRR and the NF-κB response, F. nucleatum thus orchestrates intestinal inflammation, cell-cell adhesion, autophagy fostering an environment conducive to tumor development.
Our in vitro findings unveil the significance of the ALPK1-TIFA pathway as a novel contributor to the proliferation and chemoresistance of CRC cells elicited by F. nucleatum. We show that ADP-H, released by Fusobacterium, is sufficient to recapitulate the increase in CXCL8 (IL8), ICAM1, BIRC3 and TNFAIP3 and that these effects are dependent on ALPK1/TIFA activation within IECs. Interestingly, the regulation of ULK1 was independent of this pathway, suggesting the potential involvement of other Fusobacterium-released molecules in its oncogenic effects. Moreover, the activation of the ALPK1-TIFA-NF-κB axis by ADP-H has previously been directly associated with oncogenic processes in gastric cells, in promoting replication stress and DNA damage induced by Helicobacter. Citation42 Fusobacterium has also been implicated in DNA damage in various cell-types.Citation22,Citation67–69 Therefore, we can speculate that ADP-H released by Fusobacterium may play a role in this DNA-damaging activity.
Here, we have uncovered a previously unreported phenomenon, the released ADP-H-related molecules by Fusobacterium species in their microenvironment. This release is associated with pro-oncogenic effects that do not rely on direct bacterial contact with epithelial cells. Notably the activation of the ADP-H-ALPK1-TIFA axis is independent of the contact between the bacterium and IECs, raising the possibility that it might be involved in the early stages of oncogenesis. This hypothesis presents a promising avenue for further investigation.
We have previously reported that the commensal bacterium Akkermansia muciniphila activates the ALPK1 pathway, which participates in the maintenance of intestinal barrier functions.Citation31 Our data, on both A. muciniphila and F. nucleatum point out to an ADP-H and ALPK1-dependent induction of BIRC3 and TNFAIP3 expression.Citation31 Interestingly, these genes exhibit dual functions, promoting gut homeostasis by inducing IEC proliferation and crypt regenerationCitation64,Citation65 while also being associated with tumor cell growth and survival.Citation19,Citation65 This duality emphasizes the critical importance of context in determining their effects.
In conclusion, our study has demonstrated the upregulation of oncogenic genes in CRC cells due to the F. nucleatum microenvironment, mediated by the release of ADP-H and the activation of the ALPK1-TIFA axis. These findings underscore the relevance of this pathway in CRC pathogenesis. Our results are supported by studies that have associated ALPK1 single-nucleotide polymorphisms (SNPs) and expression with various chronic inflammatory diseases and different cancers, including CRC.Citation36–40 Interestingly, recent studies have revealed links between bacteria, including Fusobacterium and a wide range of solid tumors including breast, skin, and pancreatic cancers. This suggests the existence of tumor-specific microbiota.Citation70 Therefore, our findings provide new insights into the role of tumor-associated bacteria in CRC, with potential application to other cancer types linked with ALPK1, such as lung, breast, and oral cancer.Citation71
Materials and methods
Cell culture, reporter systems and CRISPR-Cas 9 deletion
HT-29 (American Type Culture Collection, ATCC) and HEK (InvivoGen) cells were grown in RPMI1640 GlutaMAX; HeLa (ATCC) in DMEM GlutaMAX medium (Gibco), both supplemented with 10% heat-inactivated fetal bovine serum (FBS, Eurobio), with 50 IU/mL penicillin, 50 μg/mL streptomycin and 10%, 100 mM Hepes, 10 mM non-essential amino acids (Gibco). HCT116 (ATCC) cells were grown in DMEM GlutaMax with 10% heat-inactivated bovine serum (Gibco) and non-essential amino acids (Gibco). Cells were grown at 37°C in a humidified atmosphere containing 5% CO2. HT-29, HCT116 and HEK293 cells stably expressing secreted alkaline phosphatase (SEAP, pNiflty, InvivoGen) reporter gene were used to monitor NF-κB activation (HT-29-NF-κB, HCT116-NF-κB and HEK-NF-κB).Citation72 HeLa cells stably expressing TIFA-GFP were used to monitor TIFA oligomerization.Citation28 CRISPR-Cas 9 deletion of ALPK1, TIFA, MYD88 and TRAF6 in HEK293 and HT-29 cells has been previously published.Citation31
Reagents
NOD1 inhibitor (ML130, Sigma) and NOD1 ligand (IE-DAP, InvivoGen) were used at 10 µM and 10 µg/ml, respectively. ADP-heptose (ADP-H, InvivoGen) was used at concentrations ranging from 10−8 to 10−6M.
Culture of commensal bacteria, preparation of supernatants and short chain fatty acid (SCFA) concentration assessment
Bacterial strains were obtained from the DSMZ-German Collection and grown for 24 h in specific media and anaerobic culture conditions according to the Hungate methodCitation73 (Supplementary Table S1). Fusobacterium supernatants were harvested after centrifugation at 5,000 × g for 10 min, filtered through 0.22 μm PES filters, and stored at −80°C. Quality controls were performed using the Gram staining method, aerobic growth test, and fresh observations under a microscope. The concentrations of SCFAs produced by cultured bacteria were assessed by gas chromatography as described in.Citation51
Cell stimulation and reporter system assays
For each experiment, cells were seeded at 3.104 cells per well in 96-well plates for 24–48 h and then stimulated for 24 h with bacterial supernatants or non-inoculated bacterial culture medium as a control in a total culture volume of 100 μL per well prior to the reporter assay. When indicated, the cells were incubated with ML130 (10 µM), IE-DAP (10 µg/ml), or ADP-H (10−8 to 10−6M). SEAP was revealed with the Quanti-Blue reagent (InvivoGen) using a microplate reader (655 nm Infinite 200, Tecan). NF-κB activation was normalized to the control, that is, unstimulated cells. Experiments were performed in triplicate for at least three independent biological assays.
CIP and PDE treatments of F. nucleatum supernatant
F. nucleatum supernatants were treated with calf intestinal alkaline phosphatase (CIP, 100 U/ml) or phosphodiesterase (PDE, 30 mU/ml) from Crotalus adamanteus for 30 min, at 37°C as published followed by a 3 kDa-sieved filter step to eliminate the enzymes.Citation31,Citation49 The control media were treated similarly. The cells were stimulated with the treated supernatants for 24 h, and the NF-κB signal was monitored.
Cloning and expression of F. nucleatum hldA and hldC into E.Coli ΔHldE
E. coli K-12 WT and ΔHldE E. coli (JW3024) were obtained from Keio collectionCitation74 (Dharmacon). hldA from F. n. nucleatum was amplified using the following primers: (i) FhldAfor_HindIII (5’-GGGGAAGCTTAGGAGGTAAATAATGATAAGTAAATTAATAG-3’), creating a new HindIII restriction site (AAGCTT) and adding a Shine-Dalgarno sequence (AGGAGG) located six bases upstream of the start codon ATG and (ii) FhldArev_BamHI (5’-GGGGGGATCCTTAATTATTACTATATATACTGTTA-3’), creating a new BamHI restriction site (GGATCC). The 998 pb fragment was amplified using Phusion High-Fidelity DNA polymerase, A-tail with GoTaq polymerase, and cloned into the pGEM-T Easy vector (Promega Corporation) to generate pGEM-T-FhldA. After the HindIII BamHI restriction of pGEM-T-FhldA, the FhldA fragment was cloned into pGBM6 to generate the pGB-hldA vector (spc/str). Similarly, hldC from F. n. nucleatum was cloned into pBAD24 generated pBAD-FhldC vector (amp) using the following primers: (i) FhldCfor_NheI (5’-GGGGGCTAGCAGGAGGTAAATAATGGAAAGGTGGGTTTTTA-3’) and (ii) FhldCrev_HindIII (5’-GGGGAAGCTTCTATTTTTTATTAATTTTTTC-3’). At each cloning step, the insertion and fragment were verified by sequencing.
Both pGB-hldA, pBAD-hldC, and the empty vectors pBAD24 and pGBM6 were purified and electroporated into E. coli ΔHldE. Overnight bacterial cultures were washed in PBS, resuspended at OD = 1, boiled for 30 min, and stored at −20°C until use.
Real-time qPCR
HT-29 or HEK cells were seeded in 6-well culture plates at densities of 1.106 per well 24 h before stimulation. HCT116 were seeded in 12-well plates at 0.8.106 cells per well 24 h prior stimulation. Total RNA was extracted using the RNeasy mini-Kit (Qiagen) according to the manufacturer’s protocols with DNase I treatment (R&D Systems). cDNA was synthesized from 2 µg of RNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), and 100 ng was used to conduct qPCRs on ABI Prism 7700 (Applied Biosystems). The following Taqman Gene expression assay probes were used: GAPDH Hs02758991_g1, ALPK1 Hs01567926, TIFA Hs00385268, ATG7 Hs00893766, ULK1 Hs00177504, BIRC3 Hs00985029, TNFAIP3 Hs00234713, CXCL8 Hs00174103_m1. GAPDH was used for normalization. Comparisons were done with 2−ΔΔCt. Samples were tested in duplicates, at least in biological triplicates.
siRNA assay
HeLa cells seeded in 96-well plates (8000 cells/well) were transfected with 20 nM siRNA as previously described.Citation30 The cells were transfected with a validated ALPK1 siRNA (s37074, Ambion) and a non-targeting sequence (4390843, Ambion). For the ALPK1 complementation experiment, 48 h after siRNA transfection, cells were transfected with an ALPK1 cDNA construct (pCMV-ALPK1) or an empty vector (pCMV) using FuGENE 6 (Roche) prior to TIFAsome analysis, as previously described.Citation28
TIFAsome assay and quantification
HeLa TIFA-GFP cells were seeded in 96-well plates for at least one day before the experiment. Cells were washed in permeabilization buffer as previously published.Citation28 Cells were then incubated with digitonin alone as a control, digitonin plus HBP, ADP-H, medium control, or bacterial supernatant for 30 min in permeabilization buffer. To monitor TIFA oligomerization, cells were fixed after 30 min of stimulation with 4% PFA. Images were acquired using ImageXpress Micro (Molecular Devices). Image analysis and TIFAsomes quantification were performed using the custom module editor of MetaXpress as previously published.Citation28
ELISA
HT-29 cells were seeded at 1 × 106 cells per well in six well-plates 24 h prior to incubation with bacterial supernatants, non-inoculated bacterial media, or ADP-H. Supernatants were collected 24 h later and assessed by ELISA following the manufacturer’s instructions using the IL8 ELISA kit (BioLegend).
Cell proliferation assay
The percentage of viable cells was determined by MTS measurement using CellTiter 96 Aqueous One solution (Promega) according to the manufacturer’s recommendations. Briefly, the reagent solution was directly added to the culture wells, and the plates were incubated 0,5 for 1 h before measuring the absorbance at 490 nm using a microplate reader (655 nm Infinite 200, Tecan). Experiments were performed in technical triplicates for at least three independent biological assays.
Statistical analysis
All experiments were conducted in technical triplicates for at least three independent biological assays (otherwise specified), and the results are expressed as mean ± SD fold change toward the control condition. Data were represented and analyzed using GraphPad Prism (GraphPad Software). Differences between individual groups were verified using Mann-Whitney test or one-way ANOVA followed by Tukey’s multiple comparisons test. P-values are indicated as follows: ****P <.0001; ***P < .001; **P < .01; *P < .05; P >.05 was considered not significant (ns).
Authors contributions
Conceived and designed the experiments: CMG, CA, NL; intellectual contribution: JD, HMB; data acquisition: CMG, LS, DGW, LM, FBC, CLG, VB, AJ, CA, NL; data analysis, statistical analysis, and interpretation: CMG, DGW, CA, and NL; wrote the paper: CMG, NL; edited and revised the manuscript: JD, HMB, CA, DGW, and LM.
Supplemental Material
Download MS Word (13.6 KB)Supplementary figS2 resub.tiff
Download TIFF Image (1.5 MB)Supplementary fig S1 resub.tiff
Download TIFF Image (389.4 KB)Supplementary Table S1.xlsx
Download MS Excel (10.9 KB)Acknowledgments
We are grateful to Dr. Marion Espeli (INSERM U1160) for helpful discussions, critical comments, and corrections on the manuscript. The authors are grateful to Dr. Anne Letessier (Institut Cochin), Dr. Benoit Miotto (Institut Cochin) for their helpful discussion. The authors are grateful to Amandine Lashermes for technical input, Agnès David (INRAE, UMR 1280 PHAN, Nantes, France) and Camille Mayeur (INRAE Micalis) for SCFA analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data are contained in the article and the supporting information.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2023.2295384
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–19. doi:10.3322/caac.21492.
- Ananthakrishnan AN, Bernstein CN, Iliopoulos D, Macpherson A, Neurath MF, Ali RAR, Vavricka SR, Fiocchi C. Environmental triggers in IBD: a review of progress and evidence. Nat Rev Gastro Hepat. 2018;15(1):39–49. doi:10.1038/nrgastro.2017.136.
- Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, Beghini F, Manara S, Karcher N, Pozzi C, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. 2019;25(4):667–678. doi:10.1038/s41591-019-0405-7.
- Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25(4):679–689. doi:10.1038/s41591-019-0406-6.
- Allali I, Delgado S, Marron PI, Astudillo A, Yeh JJ, Ghazal H, Amzazi S, Keku T, Azcarate-Peril MA. Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microbes. 2015;6(3):161–72. doi:10.1080/19490976.2015.1039223.
- Repass J, Maherali N, Owen K. Reproducibility Project: Cancer B, Reproducibility Project Cancer B. Registered report: Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Elife. 2016;5. doi:10.7554/eLife.10012.
- Repass J, Iorns E, Denis A, Williams SR, Perfito N, Errington TM. Reproducibility Project: Cancer B. Replication Study: Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Elife. 2018;7. doi:10.7554/eLife.25801.
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy T, Chung D, Lochhead P, Hold G, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi:10.1016/j.chom.2013.07.007.
- Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi:10.1101/gr.126516.111.
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298. doi:10.1101/gr.126573.111.
- Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, Zhang D, Xia H, Xu X, Jie Z, et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat Commun. 2015;6(1):6528. doi:10.1038/ncomms7528.
- Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, Amiot A, Böhm J, Brunetti F, Habermann N, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10(11):766. doi:10.15252/msb.20145645.
- Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, Ng SC, Tsoi H, Dong Y, Zhang N, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6(1):8727. doi:10.1038/ncomms9727.
- Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, Igarashi H, Takahashi T, Tachibana M, Takahashi H, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137(6):1258–1268. doi:10.1002/ijc.29488.
- Yu J, Chen Y, Fu X, Zhou X, Peng Y, Shi L, Chen T, Wu Y. Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer. 2016;139(6):1318–26. doi:10.1002/ijc.30168.
- Dai Z, Coker OO, Nakatsu G, WKK W, Zhao L, Chen Z, Chan FKL, Kristiansen K, Sung JJY, Wong SH, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6(1):70. doi:10.1186/s40168-018-0451-2.
- Young C, Wood HM, Seshadri RA, Van Nang P, Vaccaro C, Melendez LC, Bose M, Van Doi M, Piñero TA, Valladares CT, et al. The colorectal cancer-associated faecal microbiome of developing countries resembles that of developed countries. Genome Med. 2021;13(1):27. doi:10.1186/s13073-021-00844-8.
- Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–63 e16. doi:10.1016/j.cell.2017.07.008.
- Zhang S, Yang Y, Weng W, Guo B, Cai G, Ma Y, Cai S. Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal cancer. J Exp Clin Cancer Res. 2019;38(1):14. doi:10.1186/s13046-018-0985-y.
- Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–225. doi:10.1016/j.chom.2016.07.006.
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi:10.1016/j.chom.2013.07.012.
- Guo P, Tian Z, Kong X, Yang L, Shan X, Dong B, Ding X, Jing X, Jiang C, Jiang N, et al. FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J Exp Clin Cancer Res. 2020;39(1):202. doi:10.1186/s13046-020-01677-w.
- Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi:10.1016/j.immuni.2015.01.010.
- Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear Factor−κB, and up-regulating expression of MicroRNA-21. Gastroenterology. 2017;152(4):851–66 e24. doi:10.1053/j.gastro.2016.11.018.
- Tilg H, Adolph TE, Gerner RR, Moschen AR. The intestinal microbiota in colorectal cancer. Cancer Cell. 2018;33(6):954–964. doi:10.1016/j.ccell.2018.03.004.
- Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, Huot L, Grandjean T, Bressenot A, Delanoye-Crespin A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013;123:700–711. doi:10.1172/JCI62236.
- Chen R, Alvero AB, Silasi DA, Steffensen KD, Mor G. Cancers take their toll—the function and regulation of toll-like receptors in cancer cells. Oncogene. 2008;27(2):225–33. doi:10.1038/sj.onc.1210907.
- Garcia-Weber D, Dangeard AS, Cornil J, Thai L, Rytter H, Zamyatina A, Mulard LA, Arrieumerlou C. ADP-heptose is a newly identified pathogen-associated molecular pattern of shigella flexneri. EMBO Rep. 2018;19(12). doi:10.15252/embr.201846943.
- Zhou P, She Y, Dong N, Li P, He H, Borio A, Wu Q, Lu S, Ding X, Cao Y, et al. Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose. Nature. 2018;561(7721):122–126. doi:10.1038/s41586-018-0433-3.
- Milivojevic M, Dangeard AS, Kasper CA, Tschon T, Emmenlauer M, Pique C, Schnupf P, Guignot J, Arrieumerlou C, et al. ALPK1 controls TIFA/TRAF6-dependent innate immunity against heptose-1,7-bisphosphate of gram-negative bacteria. PLoS Pathog. 2017;13(2):e1006224. doi:10.1371/journal.ppat.1006224.
- Martin-Gallausiaux C, Garcia-Weber D, Lashermes A, Larraufie P, Marinelli L, Teixeira V, Rolland A, Béguet-Crespel F, Brochard V, Quatremare T, et al. Akkermansia muciniphila upregulates genes involved in maintaining the intestinal barrier function via ADP-heptose-dependent activation of the ALPK1/TIFA pathway. Gut Microbes. 2022;14(1):2110639. doi:10.1080/19490976.2022.2110639.
- Gaudet RG, Guo CX, Molinaro R, Kottwitz H, Rohde JR, Dangeard AS, Arrieumerlou C, Girardin SE, Gray-Owen SD. Innate recognition of intracellular bacterial growth is driven by the TIFA-Dependent cytosolic surveillance pathway. Cell Rep. 2017;19(7):1418–30. doi:10.1016/j.celrep.2017.04.063.
- Cui J, Duizer C, Bouwman LI, van Rooijen KS, Voogdt CGP, van Putten JPM, de Zoete MR. The ALPK1 pathway drives the inflammatory response to Campylobacter jejuni in human intestinal epithelial cells. PLoS Pathog. 2021;17(8):e1009787. doi:10.1371/journal.ppat.1009787.
- Zhang Y, Zhang L, Zheng S, Li M, Xu C, Jia D, Qi Y, Hou T, Wang L, Wang B, et al. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-κB/ICAM1 axis. Gut Microbes. 2022;14(1):2038852. doi:10.1080/19490976.2022.2038852.
- Ryzhakov G, West NR, Franchini F, Clare S, Ilott NE, Sansom SN, Bullers SJ, Pearson C, Costain A, Vaughan-Jackson A, et al. Alpha kinase 1 controls intestinal inflammation by suppressing the IL-12/Th1 axis. Nat Commun. 2018;9(1):3797. doi:10.1038/s41467-018-06085-5.
- Strietz J, Stepputtis SS, Preca BT, Vannier C, Kim MM, Castro DJ, Au Q, Boerries M, Busch H, Aza-Blanc P, et al. ERN1 and ALPK1 inhibit differentiation of bi-potential tumor-initiating cells in human breast cancer. Oncotarget. 2016;7(50):83278–83293. doi:10.18632/oncotarget.13086.
- Liao HF, Lee HH, Chang YS, Lin CL, Liu TY, Chen YC, Yen J-C, Lee Y-T, Lin C-Y, Wu S-H, et al. Down-regulated and commonly mutated ALPK1 in lung and colorectal cancers. Sci Rep. 2016;6(1):27350. doi:10.1038/srep27350.
- Chen PK, Hua CH, Hsu HT, Kuo TM, Chung CM, Lee CP, Tsai M-H, Yeh K-T, Ko Y-C. ALPK1 expression is associated with lymph node metastasis and tumor growth in oral squamous cell carcinoma patients. Am J Pathol. 2019;189(1):190–9. doi:10.1016/j.ajpath.2018.09.003.
- Sangiorgi E, Azzara A, Molinario C, Pietrobono R, Rigante D, Verrecchia E, Sicignano LL, Genuardi M, Gurrieri F, Manna R, et al. Rare missense variants in the ALPK1 gene may predispose to periodic fever, aphthous stomatitis, pharyngitis and adenitis (PFAPA) syndrome. Eur J Hum Genet. 2019;27(9):1361–1368. doi:10.1038/s41431-019-0421-6.
- Rashid M, van der Horst M, Mentzel T, Butera F, Ferreira I, Pance A, Rütten A, Luzar B, Marusic Z, de Saint Aubain N, et al. ALPK1 hotspot mutation as a driver of human spiradenoma and spiradenocarcinoma. Nat Commun. 2019;10(1):2213. doi:10.1038/s41467-019-09979-0.
- Wei TW, Wu PY, Wu TJ, Hou HA, Chou WC, Teng CJ, Lin C-R, Chen JMM, Lin T-Y, Su H-C, et al. Aurora a and NF-κB survival pathway drive chemoresistance in acute myeloid leukemia via the TRAF-Interacting protein TIFA. Cancer Res. 2017;77(2):494–508. doi:10.1158/0008-5472.CAN-16-1004.
- Bauer M, Nascakova Z, Mihai AI, Cheng PF, Levesque MP, Lampart S, Hurwitz R, Pfannkuch L, Dobrovolna J, Jacobs M, et al. The ALPK1/TIFA/NF-κB axis links a bacterial carcinogen to R-loop-induced replication stress. Nat Commun. 2020;11(1):5117. doi:10.1038/s41467-020-18857-z.
- Wang FF, Zhao PY, He XJ, Jiang K, Wang TS, Xiao JW, Sun D-B, Guo D-H. Fusobacterium necrophorum promotes apoptosis and inflammatory cytokine production through the activation of NF-κB and death receptor signaling pathways. Front Cell Infect Microbiol. 2022;12:827750. doi:10.3389/fcimb.2022.827750.
- Martin-Gallausiaux C, Malabirade A, Habier J, Wilmes P. Fusobacterium nucleatum extracellular vesicles modulate Gut epithelial cell innate immunity via FomA and TLR2. Front Immunol. 2020;11:583644. doi:10.3389/fimmu.2020.583644.
- Chen Y, Peng Y, Yu J, Chen T, Wu Y, Shi L, Li Q, Wu J, Fu X. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget. 2017;8(19):31802–14. doi:10.18632/oncotarget.15992.
- Nomoto D, Baba Y, Liu Y, Tsutsuki H, Okadome K, Harada K, Ishimoto T, Iwatsuki M, Iwagami S, Miyamoto Y, et al. Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression via the NOD1/RIPK2/NF-κB pathway. Cancer Lett. 2022;530:59–67. doi:10.1016/j.canlet.2022.01.014.
- Alyami HM, Finoti LS, Teixeira HS, Aljefri A, Kinane DF, Benakanakere MR. Role of NOD1/NOD2 receptors in Fusobacterium nucleatum mediated NETosis. Microb Pathog. 2019;131:53–64. doi:10.1016/j.micpath.2019.03.036.
- Barnich N, Aguirre JE, Reinecker HC, Xavier R, Podolsky DK. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor–κB activation in muramyl dipeptide recognition. J Cell Biol. 2005;170(1):21–6. doi:10.1083/jcb.200502153.
- Pfannkuch L, Hurwitz R, Traulsen J, Sigulla J, Poeschke M, Matzner L, Kosma P, Schmid M, Meyer TF. ADP heptose, a novel pathogen-associated molecular pattern identified in Helicobacter pylori. FASEB J. 2019;33(8):9087–99. doi:10.1096/fj.201802555R.
- Marinelli L, Martin-Gallausiaux C, Bourhis J, Beguet-Crespel F, Blottiere HM, Lapaque N. Identification of the novel role of butyrate as AhR ligand in human intestinal epithelial cells. Sci Rep. 2019;9(1):643. doi:10.1038/s41598-018-37019-2.
- Martin-Gallausiaux C, Beguet-Crespel F, Marinelli L, Jamet A, Ledue F, Blottiere HM, Lapaque N. Butyrate produced by gut commensal bacteria activates TGF-beta1 expression through the transcription factor SP1 in human intestinal epithelial cells. Sci Rep. 2018;8(1):9742. doi:10.1038/s41598-018-28048-y.
- Martin-Gallausiaux C, Larraufie P, Jarry A, Beguet-Crespel F, Marinelli L, Ledue F, Reimann F, Blottière HM, Lapaque N. Butyrate produced by commensal bacteria down-regulates indolamine 2,3-dioxygenase 1 (IDO-1) expression via a dual mechanism in human intestinal epithelial cells. Front Immunol. 2018;9:2838. doi:10.3389/fimmu.2018.02838.
- Leung CH, Lam W, Ma DL, Gullen EA, Cheng YC. Butyrate mediates nucleotide-binding and oligomerisation domain (NOD) 2-dependent mucosal immune responses against peptidoglycan. Eur J Immunol. 2009;39(12):3529–3537. doi:10.1002/eji.200939454.
- Lakhdari O, Tap J, Beguet-Crespel F, Le Roux K, de Wouters T, Cultrone A, Nepelska M, Lefèvre F, Doré J, Blottière HM. Identification of NF-κB modulation capabilities within human intestinal commensal bacteria. J Biomed Biotechnol. 2011;2011:1–9. doi:10.1155/2011/282356.
- Larraufie P, Dore J, Lapaque N, Blottiere HM. TLR ligands and butyrate increase pyy expression through two distinct but inter-regulated pathways. Cell Microbiol. 2016;19(2):e12648. doi:10.1111/cmi.12648.
- Lucas C, Barnich N, Nguyen NH. Microbiota, Inflammation and Colorectal Cancer. Int J Mol Sci. 2017;18(6):18. doi:10.3390/ijms18061310.
- Thomas AM, Fidelle M, Routy B, Kroemer G, Wargo JA, Segata N, Zitvogel L. Gut OncoMicrobiome signatures (GOMS) as next-generation biomarkers for cancer immunotherapy. Nat Rev Clin Oncol. 2023;20(9):583–603. doi:10.1038/s41571-023-00785-8.
- Yeoh YK, Chen Z, Wong MCS, Hui M, Yu J, Ng SC, Sung JJY, Chan FKL, Chan PKS. Southern Chinese populations harbour non-nucleatum fusobacteria possessing homologues of the colorectal cancer-associated FadA virulence factor. Gut. 2020;69(11):1998–2007. doi:10.1136/gutjnl-2019-319635.
- Gaudet RG, Sintsova A, Buckwalter CM, Leung N, Cochrane A, Li J, Cox AD, Moffat J, Gray-Owen SD. INNATE IMMUNITY. Cytosolic detection of the bacterial metabolite HBP activates TIFA-dependent innate immunity. Sci. 2015;348(6240):1251–5. doi:10.1126/science.aaa4921.
- Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309–24. doi:10.1038/nri.2017.142.
- Bazzichetto C, Milella M, Zampiva I, Simionato F, Amoreo CA, Buglioni S, Pacelli C, Le Pera L, Colombo T, Bria E, et al. Interleukin-8 in colorectal cancer: a systematic review and meta-analysis of its potential role as a prognostic biomarker. Biomedicines. 2022;10(10). doi:10.3390/biomedicines10102631.
- Casasanta MA, Yoo CC, Udayasuryan B, Sanders BE, Umana A, Zhang Y, Peng H, Duncan AJ, Wang Y, Li L, et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal. 2020;13(641). doi:10.1126/scisignal.aba9157.
- Qiu Z, Wang Y, Zhang Z, Qin R, Peng Y, Tang W, Xi Y, Tian G, Zhang Y. Roles of intercellular cell adhesion molecule-1 (ICAM-1) in colorectal cancer: expression, functions, prognosis, tumorigenesis, polymorphisms and therapeutic implications. Front Oncol. 2022;12:1052672. doi:10.3389/fonc.2022.1052672.
- Vereecke L, Vieira-Silva S, Billiet T, van Es JH, Mc Guire C, Slowicka K, Sze M, van den Born M, De Hertogh G, Clevers H, et al. A20 controls intestinal homeostasis through cell-specific activities. Nat Commun. 2014;5(1):5103. doi:10.1038/ncomms6103.
- Dagenais M, Dupaul-Chicoine J, Champagne C, Skeldon A, Morizot A, Saleh M. A critical role for cellular inhibitor of protein 2 (cIAP2) in colitis-associated colorectal cancer and intestinal homeostasis mediated by the inflammasome and survival pathways. Mucosal Immunol. 2016;9(1):146–158. doi:10.1038/mi.2015.46.
- Levy J, Cacheux W, Bara MA, L’Hermitte A, Lepage P, Fraudeau M, Trentesaux C, Lemarchand J, Durand A, Crain A-M, et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol. 2015;17(8):1062–1073. doi:10.1038/ncb3206.
- Geng F, Zhang Y, Lu Z, Zhang S, Pan Y. Fusobacterium nucleatum caused DNA damage and promoted cell proliferation by the Ku70/p53 pathway in oral cancer cells. DNA Cell Biol. 2020;39(1):144–51. doi:10.1089/dna.2019.5064.
- Sayed IM, Chakraborty A, Abd El-Hafeez AA, Sharma A, Sahan AZ, Huang WJM, Sahoo D, Ghosh P, Hazra TK, Das S, et al. The DNA glycosylase NEIL2 suppresses Fusobacterium-infection-induced inflammation and DNA damage in colonic epithelial cells. Cells. 2020;9(9):1980. doi:10.3390/cells9091980.
- Okita Y, Koi M, Takeda K, Ross R, Mukherjee B, Koeppe E, Stoffel EM, Galanko JA, McCoy AN, Keku TO, et al. Fusobacterium nucleatum infection correlates with two types of microsatellite alterations in colorectal cancer and triggers DNA damage. Gut Pathog. 2020;12(1):46. doi:10.1186/s13099-020-00384-3.
- Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, et al. The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Sci. 2020;368(6494):973–80. doi:10.1126/science.aay9189.
- Ko AM, Tu HP, Ko YC. Systematic review of the role of Alpha-protein kinase 1 in cancer and cancer-related inflammatory diseases. Cancers Basel. 2022;14(18):14. doi:10.3390/cancers14184390.
- Lakhdari O, Cultrone A, Tap J, Gloux K, Bernard F, Ehrlich SD, Lefèvre F, Doré J, Blottière HM, et al. Correction: functional metagenomics: a High throughput screening method to decipher microbiota-Driven NF-κB modulation in the human Gut. PLoS ONE. 2010;5(10). doi:10.1371/annotation/9f1b7f00-bcc0-4442-9775-491ebdafc7bc.
- Hungate RE. The anaerobic mesophilic cellulolytic bacteria. Bacteriol Rev. 1950;14(1):1–49. doi:10.1128/br.14.1.1-49.1950.
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2(1):2006–0008. doi:10.1038/msb4100050.
