ABSTRACT
Melatonin has various physiological effects, such as the maintenance of circadian rhythms, anti-inflammatory functions, and regulation of intestinal barriers. The regulatory functions of melatonin in gut microbiota remodeling have also been well clarified; however, the role of gut microbiota in regulating host melatonin production remains poorly understood. To address this, we studied the contribution of gut microbiota to host melatonin production using gut microbiota-perturbed models. We demonstrated that antibiotic-treated and germ-free mice possessed diminished melatonin levels in the serum and elevated melatonin levels in the colon. The influence of the intestinal microbiota on host melatonin production was further confirmed by fecal microbiota transplantation. Notably, Lactobacillus reuteri (L. R) and Escherichia coli (E. coli) recapitulated the effects of gut microbiota on host melatonin production. Mechanistically, L. R and E. coli activated the TLR2/4/MyD88/NF-κB signaling pathway to promote expression of arylalkylamine N-acetyltransferase (AANAT, a rate-limiting enzyme for melatonin production), and MyD88 deficiency in colonic epithelial cells abolished the influence of intestinal microbiota on colonic melatonin production. Collectively, we revealed a specific underlying mechanism of gut microbiota to modulate host melatonin production, which might provide novel therapeutic ideas for melatonin-related diseases.
Introduction
Melatonin is generated from tryptophan (Trp) via the 5-hydroxytryptamine (5-HT) pathway, in which arylalkylamine N-acetyltransferase (AANAT) functions as a rate-limiting enzyme. Melatonin is produced in the pineal gland and other peripheral organs and has various physiological functions, including sleep initiation, anti-aging, and anti-inflammatory properties,Citation1–3 and its deficiency may lead to various diseases. For example, melatonin deficiency caused by sleep deprivation results in cognitive impairments and intestinal barrier dysfunction,Citation4,Citation5 and Aanat deficiency-induced melatonin reduction is involved in the pathogenesis of Alzheimer’s disease and obesity in mice.Citation6 Thus, maintaining sufficient melatonin production is crucial for health maintenance.
Melatonin synthesis is closely related to the expression of AANAT, and intrinsic mechanisms in regulation of AANAT expression have been described.Citation7,Citation8 The initiation of Aanat transcription is controlled by the norepinephrine-stimulated phosphorylation of cAMP response element-binding protein (p-CREB), as the p-CREB binds to CRE in the promoter of Aanat to initiate transcription.Citation9 Besides, extrinsic factors are also involved in melatonin production. For instance, melatonin synthesis in the pineal gland follows a clear circadian rhythm,Citation10 while melatonin production in the gastrointestinal tract could be regulated by the composition of food ingested.Citation11,Citation12 Although these factors have been well clarified, the role of gut microbiota in regulating host melatonin production remains poorly understood. Gut microbiota converts Trp into indole and its derivatives, and the role of gut microbiota in regulating the 5-HT pathway has been revealed.Citation13 Notably, gut microbiota regulates 5-HT synthesis in the serum and TPH-1 expression in the colon.Citation14,Citation15 Given that melatonin is a downstream metabolite of the Trp-5-HT pathway, we hypothesized that gut microbiota might affect host melatonin production.
Here, we found that melatonin levels decreased in the serum and increased in the colon of metronidazole-treated and germ-free (GF) mice. The influence of intestinal microbiota on host melatonin production was further confirmed by fecal microbiota transplantation. Notably, Lactobacillus reuteri (L. R) and Escherichia coli (E. coli) recapitulated the effects of gut microbiota on host melatonin production. Mechanistically, L. R and E. coli activated the TLR2/4/MyD88/NF-κB signaling pathway to promote AANAT expression, which supports melatonin production in the colon. MyD88 deficiency in the colonic epithelial cells abolished the influence of intestinal microbiota on colonic melatonin production. Overall, we revealed the underlying mechanism of gut microbiota in regulating host melatonin production, which might provide insights for melatonin-based therapeutics.
Results
Gut microbiota is involved in host melatonin production
Gut microbiota can influence host Trp metabolism,Citation16 but whether gut microbiota affects host melatonin production is unknown. In order to deplete gut microbiota and exclude the specificity of antibiotics, we selected different kinds of broad-spectrum antibiotics (ampicillin, colistin, metronidazole, neomycin, and vancomycin) for gut microbiota depletion according to previous studies.Citation17,Citation18 We treated mice with these antibiotics and measured levels of metabolites in the Trp-5-HT pathway, including Trp, 5-hydroxytryptophan (5-HTP), 5-HT, N-acetylserotonin (NAS), and melatonin by targeted metabolomics (). The metabolism of Trp-5-HT pathway was affected in mice treated with different antibiotics. Notably, ampicillin and metronidazole decreased serum melatonin levels (), whereas ampicillin, colistin, metronidazole, and neomycin increased colonic melatonin levels (). However, no significant change in melatonin levels was observed in the feces of metronidazole-treated mice (Figure S1e). These results suggest that antibiotic administrations result in diverse melatonin changes in mouse serum and colon.
Figure 1. Gut microbiota is involved in host melatonin production.
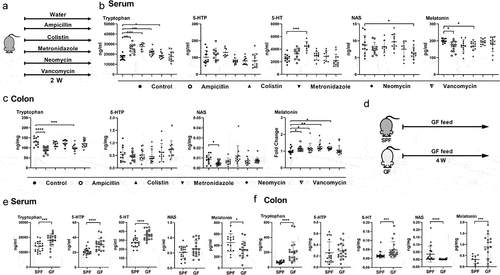
To further validate the effects of gut microbiota on melatonin levels, we measured levels of metabolites in the Trp-5-HT pathway in germ-free (GF) mice (). Similar to mice treated with antibiotics, metabolism of Trp-5-HT pathway was also affected in the serum and colon of GF mice, like lower level of serum melatonin and higher level of colonic melatonin (). Collectively, these results suggest that melatonin production is altered in the serum and colon of GF mice.
Melatonin production is regulated by fecal microbiota transplantation
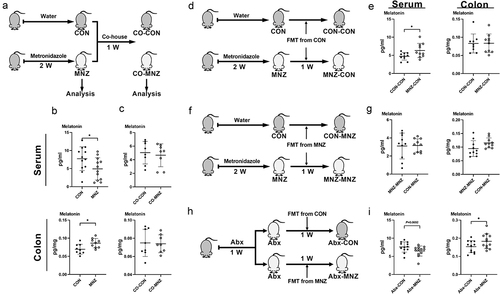
To further validate the influence of gut microbiota on melatonin production, we co-housed mice that were pretreated and not pretreated with metronidazole (). Although metronidazole administration decreased serum melatonin and elevated colonic melatonin (), the two groups of co-housed mice showed comparable melatonin levels in both the serum and colon (). We then investigated whether fecal microbiota transplantation (FMT) could alter melatonin levels in mice. FMT from the control group resulted in increased serum melatonin levels in mice pretreated with metronidazole, as well as comparable colonic melatonin levels between the control and metronidazole-pretreated groups of mice (). After receiving FMT from the metronidazole-pretreated group, alterations in melatonin levels in the serum or colon in both groups of mice that were previously detected were eliminated (). Notably, mice that were pretreated with a cocktail of antibiotics (Abx) received FMT from control mice and metronidazole-treated mice, respectively, and showed similar patterns of melatonin level alterations as their donor mice (). Overall, these results suggest that melatonin alterations in the serum and colon of mice is regulated by intestinal microbiota.
Figure 2. Melatonin production is regulated by fecal microbiota transplantation.
Lactobacillus reuteri and Escherichia coli are involved in host melatonin production
To characterize how metronidazole-induced alterations in bacteria are related to melatonin production in mice, we performed 16S rDNA sequencing of fecal bacterial DNA collected from metronidazole-treated mice to obtain microbial species richness. In LEfSe analysis, various bacterial species showed changes in mice treated with metronidazole (Figure S2a). Next, Spearman’s rank correlation was performed to characterize the relationship between microbial species richness and melatonin levels in the serum and colon, respectively. Various bacterial species were associated with serum and colonic melatonin levels, among which Lactobacilli including Lactobacillus murinus (L. M), Lactobacillus intestinalis (L. I), Lactobacillus johnsonii (L. J) and Lactobacillus reuteri (L. R), were positively correlated with serum melatonin levels but negatively correlated with colonic melatonin levels (Figure S2b-c). The abundance of E. coli was negatively correlated with serum melatonin levels (Figure S2B). Notably, the abundance of L. M, L. I, L. J and L. R were decreased but E. coli was increased in mice treated with metronidazole (Figure S2d-e). Previous studies have shown that Lactobacilli promotes melatonin production, and Lactobacillus rhamnosus enhances the abundance of melatonin receptors,Citation19,Citation20 suggesting that Lactobacilli may regulate host melatonin production. In addition, a correlation between E. coli and melatonin production in host cells has also observed.Citation21–23 These results suggest that L. M, L. I, L. J, L. R or E. coli may regulate host melatonin production.
To further determine their roles in modulating melatonin production, strains of L. M, L. R, L. I, L. J or E. coli were individually colonized into our previous established pseudo-GF (antibiotic cocktail pretreated) model, in which an antibiotic cocktail containing ampicillin, metronidazole, neomycin, and vancomycin was used for maximal microbiome depletion.Citation24,Citation25 In antibiotic cocktail-pretreated mice, L. R colonization decreased serum melatonin levels and increased colonic melatonin levels (). Although had little effect on serum melatonin levels in the control group, L. I colonization decreased serum melatonin levels in the Abx cocktail-pretreated group (). In addition, L. J colonization decreased serum melatonin levels in Abx cocktail-pretreated mice (). In order to further verify the effects of L. R on host melatonin production, L. R was colonized into GF mice. The relative abundance of Lactobacillus in both the small and large intestines of GF mice was much higher than that in the control group (undetected), and L. R colonization decreased serum melatonin levels (). However, the change in melatonin production induced by L. R, L. I or L. J colonization was inconsistent with the above spearman’s rank correlation analysis (Figure S2b-c), which may be related to the regulation of post-antibiotic gut mucosal composition, function and microbiome reconstitution by these strains.Citation26,Citation27 In Abx cocktail-pretreated mice, E. coli colonization decreased serum melatonin levels and increased colonic melatonin levels (), whereas L. M colonization did not affect host melatonin production (Figure S3a-c), indicating the strain specific regulatory function in melatonin production. Collectively, these results suggest that L. R, L. I, L. J or E. coli colonization affects host melatonin production in the serum and colon.
Figure 3. L. R, L. I, L. J or E. coli colonization affects melatonin production in vivo.
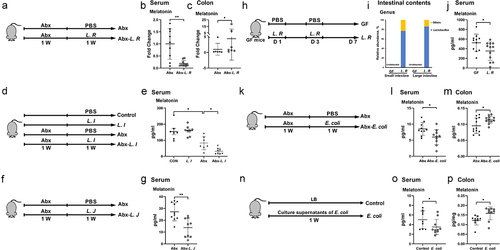
To explore the underlying molecular mechanisms of above commensal bacterial strains in host melatonin production, we cultured these bacterial strains in vitro and detected melatonin levels in their respective culture supernatants. Melatonin was not detected in any culture supernatant at any time point (Figure S4a-f), suggesting that alterations in melatonin levels we previously detected might be a result of interaction between gut microbes and the host. Compelling results indicate links between microbial metabolites and host metabolism,Citation28,Citation29 therefore, we hypothesized that these bacteria affect host melatonin production through their metabolites. Given that L. R, L. I and L. J all belong to the same bacterial taxa and have similar effects on serum melatonin production, we assumed that the common metabolite, lactate, may contribute to the effects of L. R, L. I or L. J on melatonin production. To test this hypothesis, we orally administrated culture supernatants of L. R, L. I or L. J or lactate to mice. Unlike what was observed in L. R, L. I and L. J colonized mice, all culture supernatants or lactate administration failed to recapitulate the effects of L. R, L. I or L. J colonization on serum melatonin in mice, whereas administration of L. R culture supernatants to mice decreased colonic melatonin levels (Figure S5a-c). Interestingly, the administration of E. coli culture supernatants recapitulated the effect of E. coli colonization in mice, which decreased serum melatonin levels and increased colonic melatonin levels (). Thus, the underlying molecular mechanisms of above commensal bacterial strains in host melatonin production need further well-designed investigations.
Gut microbiota promotes AANAT expression through NF-κB signaling
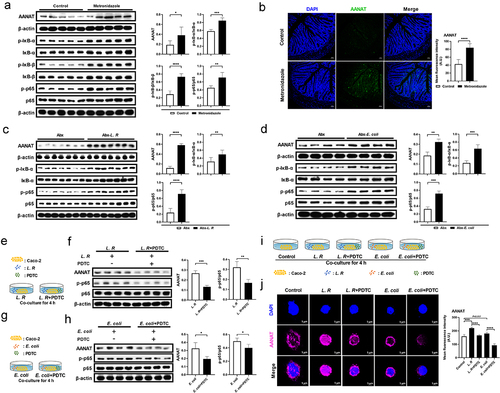
AANAT is a rate-limiting enzyme for melatonin biosynthesis and exhibits alteration synchronous with melatonin level. Therefore, in this study, we detected AANAT expression in mouse models with elevated colonic melatonin levels (). Immunoblotting results showed that metronidazole administration up-regulated AANAT expression and activated nuclear factor kappa-B (NF-κB) signaling in mouse colon (). Immunofluorescence staining also confirmed that metronidazole administration elevated AANAT expression in the colon, particularly in colonic epithelial cells (). Consistently, elevated AANAT expression and NF-κB activation were observed in mice with L. R or E. coli colonization (). Subsequently, in order to verify whether the elevation of AANAT expression resulting from gut microbiota alterations depends on NF-κB signaling, Caco-2 cells that were co-cultured with L. R or E. coli were treated with a NF-κB inhibitor, pyrrolidine dithiocarbamate ammonium (PDTC) (). As expected, the inhibition of NF-κB signaling blocked the elevation of AANAT expression in Caco-2 cells co-cultured with L. R or E. coli (). These results indicate that intestinal microbiota promotes AANAT expression via NF-κB signaling.
Figure 4. Gut microbiota regulates AANAT expression through NF-κB signaling.
Myd88 deficiency in intestinal epithelial cells blocks the effects of gut microbiota on host melatonin production
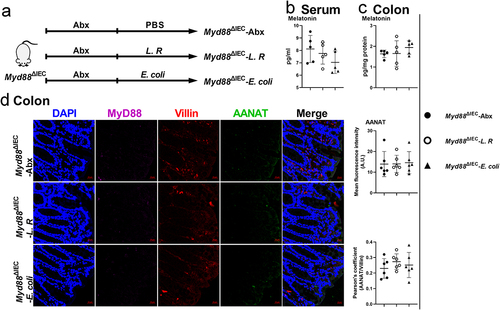
We then investigated how gut microbiota activates NF-κB signaling in colonic epithelial cells. Given that gut microbiota activates NF-κB signaling, which functions downstream of MyD88, we crossed Myd88flox/flox mice with Villin-Cre mice to generate intestinal epithelial Myd88 depleted (Myd88ΔIEC) mice (Figure S6a-b). Notably, there was no difference in serum or colonic melatonin levels observed in Myd88ΔIEC mice administered with metronidazole, which was not the case for Myd88flox/flox mice (). However, we still observed an increased abundance of MyD88, AANAT, and the ratio of p-p65/p65 in the colon of metronidazole-treated Myd88ΔIEC mice (), which might be due to the use of entire colonic tissue in immunoblotting. To rule out the involvement of other cell types, we colocalized MyD88 and AANAT with Villin (a marker of intestinal epithelial cells) and found Myd88ΔIEC mice, regardless of whether metronidazole treated, showed a comparable abundance of MyD88 and AANAT, which were Villin colocalized in colonic epithelial cells (). These results suggest that colonic epithelial cell MyD88 signaling is required for AANAT elevation induced by metronidazole treatment. MyD88 deficiency in mouse colonic epithelial cells eliminated serum and colonic melatonin level alterations induced by L. R or E. coli colonization (). In addition, L. R or E. coli colonization had no effect on expression of AANAT colocalized with Villin in Myd88ΔIEC mice (). Taken together, MyD88 deficiency in colonic epithelial cells blocks the effects of gut microbiota on host melatonin production.
Figure 5. MyD88 deficiency eliminates the effects of metronidazole treatment on melatonin production.
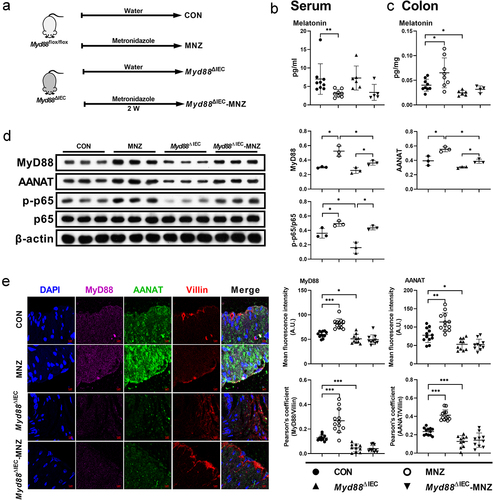
Figure 6. MyD88 deficiency eliminates the effects of L. R or E. coli colonization on melatonin production.
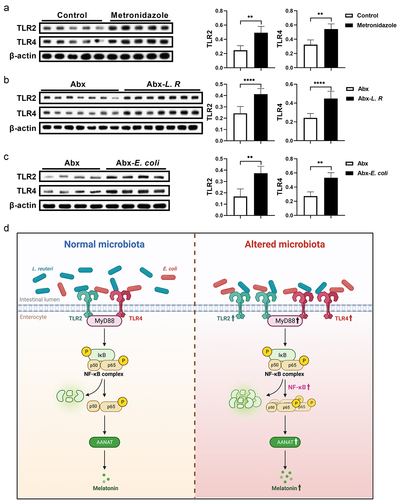
Gut microbiota activates the NF-κB pathway through TLR2 and TLR4 signaling
Given that MyD88 adapter proteins link members of the toll-like receptors (TLRs), such as TLR2 and TLR4, to activate NF-κB,Citation30 we analyzed abundance of TLR2 and TLR4 in mouse colon with altered melatonin levels (). Interestingly, both TLR2 and TLR4 were significantly increased in the colon of metronidazole-treated and L. R/E. coli colonized mice (). Taken together, these results revealed that gut microbiota regulates host melatonin production via TLR2 and TLR4 signaling, which further elevates AANAT expression by activating the MyD88/NF-κB signaling pathway ().
Figure 7. Gut microbiota activates TLR2 and TLR4 signaling in the colon.
Discussion
Melatonin exhibits antioxidative, anti-inflammatory, and anti-infective properties by remodeling gut microbiomes.Citation2,Citation3,Citation24 However, little attention has been paid to the regulation of gut microbiota on host melatonin production. In this study, we observed decreased levels of serum melatonin and increased levels of colonic melatonin in metronidazole-treated and GF mice, suggesting that gut microbiota regulates host melatonin production. Interestingly, the gut microbiota induced changes of melatonin levels are different in the serum and colon. It is known that melatonin in the blood is mainly synthesized and released by the pineal gland, while peripheral organs, including gastrointestinal tract, retina and skin, can also synthesize melatonin.Citation31 The secretion of melatonin in the pineal gland is inhibited by NF-κB signaling,Citation32 while activation of NF-κB signaling stimulates immunocompetent cells to promote melatonin production,Citation23 suggesting that the opposite change of melatonin levels in the serum and colon may be due to the bidirectional regulation of NF-κB signaling on pineal gland and colon. However, the underlying mechanism for this difference needs more well-designed investigations. Notably, we only investigated serum and colonic melatonin level alterations caused by gut microbiota clearance, whether gut microbiota regulates melatonin production in other tissues requires further exploration.
Gut microbiota is associated with host metabolism of energy, lipids, and glucose.Citation33 For instance, Klebsiella pneumoniae sequence type 258 (Kp ST258) induces host glutaminolysis and fatty acid oxidation,Citation34 Bifidobacterium longum_MG723 or Bifidobacterium bifidum_MG731 improves glucose homeostasis in diet-induced obese mice,Citation35 and Clostridium difficile promotes the production of sorbitol.Citation36 Here, L. R or E. coli colonization results in decreased melatonin levels in the serum and increased melatonin levels in the colon of Abx cocktail pretreated mice. Although we observed altered melatonin levels in mice treated with ampicillin, we did not identify bacteria that could potentially regulate melatonin production. Since antibacterial spectra are different for antibiotics, other bacteria in the gut might also affect host melatonin production. For instance, Roseburia hominis increases intestinal melatonin levels via its metabolites, propionate and butyrate.Citation37 Furthermore, we only used antibiotics to alter the composition of intestinal bacteria; the roles of fungi and viruses in host metabolism regulation, however, cannot be ignored,Citation38,Citation39 and whether they are also involved in the regulation of host melatonin production requires further investigation. Therefore, fully exploring and evaluating the potential effects of gut microbiota on host melatonin production might be a potential direction for the targeted regulation of melatonin homeostasis.
Microbial metabolites, such as short-chain fatty acids (SCFAs), indole-3-pyruvic acid (IPA), and trimethylamine (TMA), can be sensed by the intestinal epithelium and orchestrate host physiology or diseases.Citation40,Citation41 For example, Akkermansia muciniphila ameliorates metabolic disorders by regulating genes related to intestinal barrier function via TLRs.Citation42–44 In this study, administration of E. coli culture supernatants decreases the level of serum melatonin and elevates its colonic level, which is similar to the effect of E. coli colonization, but whether they function through the same mechanism is still unclear. Interestingly, we observed that L. R colonization increases colonic melatonin levels in mice, while oral administration of L. R culture supernatants decreases colonic melatonin levels in mice. Thus, oral administration of L. R culture supernatants may have different mechanisms to regulate colonic melatonin production, compared to L. R colonization. After the colonization of L. R, the composition of intestinal microbiota is changed, in which L. R occupies an important niche and increases colonic melatonin levels by activating the TLR2/4/MyD88/NF-κB signaling pathway. It is known that bacterial culture supernatants contain bacterial metabolites which affect the microbial community. For example, the culture supernatants of Clostridium difficile impact common gut microbiota associated with colonization resistance,Citation45 and the culture supernatants of Bacteroides directly limits the growth of Salmonella typhimurium.Citation46 However, the mechanism by which L. R supernatants regulate intestinal microbial composition and colonic melatonin production needs further investigation. Indeed, gut microbiota also influences host metabolism through bacterial derivatives such as microRNA, extracellular vesicles, and membrane proteins.Citation47,Citation48 For example, gut microbial DNA-containing extracellular vesicles (mEVs) are delivered to the metabolic tissues in obesity, and the leakage of mEVs contributes to the development of obesity-associated inflammation and metabolic diseases.Citation49 Unfortunately, we did not reveal how L. R activates TLR2/4/MyD88/NF-κB signaling, but the potential of L. R as a probiotic in regulating melatonin production is worthy of further exploration.
Melatonin is involved in the pathogenesis of many diseases.Citation50–52 Importantly, melatonin has an anti-inflammatory effect by affecting functions of immune cells, such as T cells, B cells, and macrophages.Citation53–56 For example, melatonin blocks the differentiation of pathogenic Th17 cells and boosts the generation of protective Tr1 cells in mice with multiple sclerosis.Citation57 Moreover, melatonin shows anti-infective activities in inhibiting Gram-negative pathogens by targeting citrate synthaseCitation50,Citation58 and fighting against coronaviruses in vitro at pharmacological concentrations.Citation59 Mechanisms through which melatonin exhibits anti-cancer effects have been well explained, including the inhibition of proliferation, invasion, metastasis, and angiogenesis, as well as the promotion of apoptosis and cancer immunity.Citation60 However, studies on melatonin therapies mostly focus on the impact of oral melatonin administration on diseases, more attention should be paid to the regulation of autonomous synthesis of melatonin.
Conclusions
In conclusion, the present results reveal the regulatory role of gut microbiota in host melatonin production. Mechanistically, gut microbiota activates TLR2/4/MyD88/NF-κB signaling in intestinal epithelial cells, resulting in higher AANAT expression and melatonin levels in the colon. This mechanism may provide novel insights for the regulation of autonomous synthesis of melatonin.
Methods
Mice
Germ-free (GF) mice were kindly provided by Prof. Hong Wei, from Huazhong Agricultural University (Wuhan, China). GF and specific-pathogen-free (SPF) BALB/c mice were bred at the Third Military Medical University (Chongqing, China). GF Kunming (KM) mice for L. R colonization were bred at the Laboratory Animal Center of Huazhong Agricultural University (Wuhan, China). Female ICR mice were purchased from SLAC Laboratory Animal Center (Changsha, China). C57BL/6J Myd88flox/flox Villin-Cre mice were kindly provided by Prof. Bie Tan, from Hunan Agricultural University (Changsha, China). In all experiments, 6 to 8 weeks female mice were used and the mice in different groups were individually housed under a 12:12 light: dark cycle unless stated otherwise. All mouse protocols were approved by the conduction of the Laboratory Animal Ethical Commission of South China Agricultural University (Guangzhou, China).
GF BALB/c mice were housed in autoclaved sterile microisolators with positively pressured HEPA-filtered sterile air, irradiated food, and autoclaved water. SPF BALB/c mice were kept in a normal environment.
For antibiotic treatment in , ICR mice were treated with ampicillin (1 g/L), metronidazole (1 g/L), colistin (1 g/L), neomycin (1 g/L), or vancomycin (0.5 g/L) in drinking water for two weeks. For metronidazole treatment in , Myd88ΔIEC mice were also treated with metronidazole (1 g/L) in drinking water for two weeks.
For co-housing, ICR mice pretreated with metronidazole for two weeks or not were co-housed in one cage for one week. Normal drinking water was provided during the co-housing experiments.
For fecal microbiota transplantation (FMT), recipient ICR mice were pretreated with metronidazole for two weeks or not, or pretreated with a cocktail of antibiotics containing neomycin (0.5 g/L), ampicillin (1 g/L), metronidazole (1 g/L), and vancomycin (0.5 g/L) for one week. FMT was then conducted for one week. Fecal material was collected from mice pretreated with metronidazole or not, resuspended at 100 mg/mL in sterile PBS, and administered via oral gavage (200 µL/mouse).
For L. M, L. R, L. J or E. coli colonization, recipient ICR mice were pretreated with Abx cocktail for one week and discontinued one day before bacteria colonization, and oral gavage with 200 μL of bacteria suspension (2 × 108 CFU/day) for one week. Especially, GF KM mice received oral gavage with 200 μL of L. R suspension (2 × 108 CFU/day) twice a week. For L. I colonization, ICR mice pretreated with Abx cocktail or not received oral gavage with 200 μL of L. I suspension (2 × 10Citation8 CFU/day) for one week. For L. R or E. coli colonization in Myd88ΔIEC mice, mice were pretreated with Abx cocktail for one week and discontinued for one day, followed by oral gavage with 200 μL of bacteria suspension (2 × 10Citation8 CFU/day) for one week.
For bacterial culture supernatant supplement in ICR mice, L. R, L. I, L. J and E. coli were cultured at 37°C for 24 h and the culture supernatants were collected. Then mice were received the supernatants by oral gavage (200 μL/day) for one week. For E. coli culture supernatant gavage, mice were administrated with LB broth as the control group. For L. R, L. I and L. J culture supernatant gavage, the control group was administrated with MRS broth, and the lactate group was orally administrated with lactate (2.6 mol/L).
Bacteria
In this study, Lactobacillus murinus (L. M), Lactobacillus reuteri (L. R), Lactobacillus johnsonii (L. J), and Escherichia coli (E. coli) strains were isolated from fecal samples of ICR mice. Lactobacillus intestinalis (L. I) was provided by Prof. Zhihong Sun, from Inner Mongolia Agricultural University (Inner Mongolia, China). L. R, L. I and L. J were cultured in MRS broth (Qingdao Rishui Bio-Technologies Co., Ltd.), E. coli was cultured in LB broth (Sangon Biotech).
Caco-2 cells
Caco-2 cells were cultured in RPMI 1640 supplemented with 10% FBS at 37°C with 5% CO2. Caco-2 cells (1 × 106 cells per well) were plated in 6-well plates and were cocultured with live L. R (1 × 10Citation8 or E. coli (1 × 108 for 4 h. The NF-κB inhibitor pyrrolidine dithiocarbamate ammonium (PDTC, 50 μM, MedChemExpress) was added with L. R or E. coli.
Elisa
The melatonin measurements of serum and colon samples were performed using a Mouse Melatonin ELISA Kit (Mlbio) in , , , , and Figure S5c. The concentrations of melatonin in the serum or colon samples were quantified using the ELISA kit according to the manufacturer’s protocol.
Immunofluorescence staining
The primary antibodies, AANAT (17990–1-AP) was purchased from Proteintech, and MyD88 (ab28763) and Villin (ab201989) were purchased from Abcam. Immunofluorescence was performed according to our previous protocols.Citation61
Western blotting
The primary antibodies AANAT (17990–1-AP), IκB-α (10268–1-AP), IκB-β (12660–1-AP), p65 (10745–1-AP), MyD88 (23230–1-AP), TLR2 (66645–1-Ig), TLR4 (19811–1-AP), and β-actin (66009–1-Ig) were purchased from Proteintech. p-IκB (ab133462) and p-p65 (ab76302) were purchased from Abcam. p-IκB-β (#4921) was purchased from Cell Signaling Technology. Colonic tissue (25 mg) was added 300 μL radio immunoprecipitation assay (RIPA) lysis buffer (Beyotime Biotechnology) for grinding and held on ice for 10 min. The supernatants were collected after centrifugation for Western blotting. Western blotting assays of Caco-2 cells were carried out using routine procedures.
UPLC-Orbitrap-MS/MS conditions
The levels of Trp metabolites in the serum, colon and feces of mice and bacterial supernatants were analyzed using UPLC-Orbitrap-MS/MS. The conditions have been described in our previous study.Citation62 Briefly, melatonin and other Trp metabolites were separated using a Thermo Fisher Scientific UPLC system (Dionex UltiMate 3000) and a C1s Hypersil Gold column (1.9 μm, 100 mm × 2.1 mm; Thermo Scientific).
16S rRNA gene sequencing analysis
16S rRNA gene sequencing and analysis were conducted by a commercial company (Novogene, Co., Ltd.). Briefly, DNA form intestinal content was extracted with QIAamp DNA Stool Mini Kit (Qiagen) following manufacturer’s instructions. Ribosomal 16S rDNA V4 region was amplified with universal primers: 515 F (5’-GTGCCAGCMGCCGCGGTAA-3’) and 806 R (5’-GGACTACHVGGGTWTCTAAT-3’). PCR reactions were carried out with 15 µL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs), 0.2 µM of forward and reverse primers, and 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and elongation at 72°C for 30 s and 72°C for 5 min. Then, PCR products were purified with Universal DNA Purification Kit (TianGen). Sequencing libraries were generated using NEB Next® Ultra™ II FS DNA PCR-free Library Prep Kit (New England Biolabs) following manufacturer’s recommendations. The library was checked with Qubit and real-time PCR for quantification and bioanalyzer for size distribution detection. Quantified libraries were pooled and sequenced on Illumina platforms according to effective library concentration. Paired-end reads were assigned to samples based on their unique barcode and truncated by cutting off the barcode and primer sequence. Paired-end reads were merged using FLASH (V1.2.11, http://ccb.jhu.edu/software/FLASH/), and the splicing sequences were called raw tags. Quality filtering on the raw tags were performed using the fastp (Version 0.23.1) software to obtain high-quality Clean Tags. The tags were compared with the reference database (Silva database (16S/18S), https://www.arb-silva.de/; Unite Database (ITS), https://unite.ut.ee/) using UCHIME Algorithm (http://www.drive5.com/usearch/manual/uchime_algo.html) to detect chimera sequences, and then the effective tags were finally obtained. The effective tags were analyzed (by using Uparse software, version 7.0.1001), and assigned to the same OTUs with ≥ 97% similarity. For further annotation, we screened the representative sequence from each OTU. Species annotation was performed on OTUs sequences using the Mothur method and the SSUrRNA database of SILVA138 (http://www.arb-silva.de/) (with a threshold of 0.8–1). Taxonomic information was obtained, and the community composition of each sample was analyzed at various taxonomic levels of kingdom, phylum, class, order, family, genus, and species.
Statistical analysis
For target metabolomic analysis, data in an instrument-specific format (*raw) were converted by the Xcalibur (version 3.0, Thermo Fisher) to common data format (XLS) files, which show information on metabolites, including the calculated amount, retention time, and peak areas. Melatonin levels in different experiments were normalized to those in their control groups. Data and statistical plots in this study were processed by GraphPad Prism 8.0 (GraphPad Software). All data are presented as mean ± standard deviation (SD). Statistical analysis was conducted as described previously.Citation24 Data between two groups were analyzed by unpaired t test if the data are in Gaussian distribution and have equal variance, or by unpaired t test with Welch’s correction if the data are in Gaussian distribution but show unequal variance, or by non-parametric test (Mann – Whitney U test) if the data are not normally distributed. Multigroup were analyzed by one-way ANOVA followed by Dunnett multiple comparisons if the date were in Normal distribution and had equal variance, or else calculated by Kruskal-Wallis followed by Dunnett multiple comparisons. p < 0.05 was defined as significant difference. The data in figures is presented as *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.
Authors’ contributions
W.K.R. supervised the entire project and designed the experiments. B.N.L. and L.J.F. performed the experiments and analyzed the data. Y.X.W., H.W., Y.Q.Y., S.C., I.F.H., C.X.L., H.W., and L.P.G. assisted with experiments. B.N.L. wrote the paper. W.K.R. revised the manuscript. All the authors have read and approved the final version of the manuscript.
Supplemental Material
Download Zip (3 MB)Acknowledgments
We thank Prof. Bie Tan (Hunan Agricultural University, Changsha, China) for kindly support of Myd88flox/flox Villin-Cre mice.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The raw 16S rDNA sequencing data generated in this study have been deposited in the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under the accession number PRJNA1047195.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2024.2313769
Additional information
Funding
References
- Boutin JA, Jockers R. Melatonin controversies, an update. J Pineal Res. 2021;70:e12702. doi:10.1111/jpi.12702.
- Yi X, Cai R, Shaoyong W, Wang G, Yan W, He Z, Li R, Chao M, Zhao T, Deng L. et al. Melatonin promotes gut anti-oxidative status in perinatal rat by remodeling the gut microbiome. Redox Biol. 2023;65:102829. doi:10.1016/j.redox.2023.102829.
- Liu S, Kang W, Mao X, Ge L, Du H, Li J, Hou L, Liu D, Yin Y, Liu Y. et al. Melatonin mitigates aflatoxin B1-induced liver injury via modulation of gut microbiota/intestinal FXR/liver TLR4 signaling axis in mice. J Pineal Res. 2022;73(2):e12812. doi:10.1111/jpi.12812.
- Gao T, Wang Z, Dong Y, Cao J, Lin R, Wang X, Yu Z, Chen Y. Role of melatonin in sleep deprivation-induced intestinal barrier dysfunction in mice. J Pineal Res. 2019;67(1):e12574. doi:10.1111/jpi.12574.
- Wang X, Wang Z, Cao J, Dong Y, Chen Y. Gut microbiota-derived metabolites mediate the neuroprotective effect of melatonin in cognitive impairment induced by sleep deprivation. Microbiome. 2023;11(1):17. doi:10.1186/s40168-022-01452-3.
- Zhang B, Chen T, Cao M, Yuan C, Reiter RJ, Zhao Z, Zhao Y, Chen L, Fan W, Wang X. et al. Gut microbiota dysbiosis induced by decreasing endogenous melatonin mediates the pathogenesis of Alzheimer’s disease and obesity. Front Immunol. 2022;13:900132. doi:10.3389/fimmu.2022.900132.
- Villela D, de Sá Lima L, Peres R, Peliciari-Garcia RA, Do Amaral FG, Cipolla-Neto J, Scavone C, Afeche SC. Norepinephrine activates NF-κB transcription factor in cultured rat pineal gland. Life Sci. 2014;94:122–17. doi:10.1016/j.lfs.2013.11.004.
- Koch M, Mauhin V, Stehle JH, Schomerus C, Korf HW. Dephosphorylation of pCREB by protein serine/threonine phosphatases is involved in inactivation of aanat gene transcription in rat pineal gland. J Neurochem. 2003;85(1):170–179. doi:10.1046/j.1471-4159.2003.01651.x.
- Ho AK, Chik CL. Modulation of aanat gene transcription in the rat pineal gland. J Neurochem. 2010;112:321–331. doi:10.1111/j.1471-4159.2009.06457.x.
- Foster RG. Melatonin. Curr Biol. 2021;31(22):R1456–r1458. doi:10.1016/j.cub.2021.10.029.
- Yasmin F, Sutradhar S, Das P, Mukherjee S. Gut melatonin: a potent candidate in the diversified journey of melatonin research. Gen Comp Endocr. 2021;303:113693. doi:10.1016/j.ygcen.2020.113693.
- Bubenik GA, Pang SF, Hacker RR, Smith PS. Melatonin concentrations in serum and tissues of porcine gastrointestinal tract and their relationship to the intake and passage of food. J Pineal Res. 1996;21(4):251–256. doi:10.1111/j.1600-079x.1996.tb00294.x.
- Zhu P, Lu T, Wu J, Fan D, Liu B, Zhu X, Guo H, Du Y, Liu F, Tian Y. et al. Gut microbiota drives macrophage-dependent self-renewal of intestinal stem cells via niche enteric serotonergic neurons. Cell Res. 2022;32(6):555–569. doi:10.1038/s41422-022-00645-7.
- Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161(2):264–276. doi:10.1016/j.cell.2015.02.047.
- Reigstad CS, Salmonson CE, Rainey JF 3rd, Szurszewski JH, Linden DR, Sonnenburg JL, Farrugia G, Kashyap PC. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015;29(4):1395–1403. doi:10.1096/fj.14-259598.
- Han JX, Tao ZH, Wang JL, Zhang L, Yu CY, Kang ZR, Xie Y, Li J, Lu S, Cui Y. et al. Microbiota-derived tryptophan catabolites mediate the chemopreventive effects of statins on colorectal cancer. Nat Microbiol. 2023;8(5):919–933. doi:10.1038/s41564-023-01363-5.
- Miyauchi E, Kim SW, Suda W, Kawasumi M, Onawa S, Taguchi-Atarashi N, Morita H, Taylor TD, Hattori M, Ohno H. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature. 2020;585(7823):102–106. doi:10.1038/s41586-020-2634-9.
- Reeves AE, Theriot CM, Bergin IL, Huffnagle GB, Schloss PD, Young VB. The interplay between microbiome dynamics and pathogen dynamics in a murine model of clostridium difficile infection. Gut Microbes. 2011;2(3):145–158. doi:10.4161/gmic.2.3.16333.
- Tan DX, Hardeland R, Manchester LC, Rosales-Corral S, Coto-Montes A, Boga JA, Reiter RJ. Emergence of naturally occurring melatonin isomers and their proposed nomenclature. J Pineal Res. 2012;53(2):113–121. doi:10.1111/j.1600-079X.2012.00979.x.
- Lutfi E, Basili D, Falcinelli S, Morillas L, Carnevali O, Capilla E, Navarro I. The probiotic Lactobacillus rhamnosus mimics the dark-driven regulation of appetite markers and melatonin receptors’ expression in zebrafish (danio rerio) larvae: understanding the role of the gut microbiome. Comp Biochem Physiol Part B: Biochem Mol Biol. 2021;256:110634. doi:10.1016/j.cbpb.2021.110634.
- Paulose JK, Wright JM, Patel AG, Cassone VM. Human gut bacteria are sensitive to melatonin and express endogenous circadian rhythmicity. PloS ONE. 2016;11(1):e0146643. doi:10.1371/journal.pone.0146643.
- Lopes JG, Sourjik V. Chemotaxis of Escherichia coli to major hormones and polyamines present in human gut. The ISME Journal. 2018;12(11):2736–2747. doi:10.1038/s41396-018-0227-5.
- Pontes GN, Cardoso EC, Carneiro-Sampaio MM, Markus RP. Injury switches melatonin production source from endocrine (pineal) to paracrine (phagocytes) – melatonin in human colostrum and colostrum phagocytes. J Pineal Res. 2006;41(2):136–141. doi:10.1111/j.1600-079X.2006.00345.x.
- Ren W, Wang P, Yan J, Liu G, Zeng B, Hussain T, Peng C, Yin J, Li T, Wei H. et al. Melatonin alleviates weanling stress in mice: involvement of intestinal microbiota. J Pineal Res. 2018;64(2):64. doi:10.1111/jpi.12448.
- Yin J, Li Y, Han H, Chen S, Gao J, Liu G, Wu X, Deng J, Yu Q, Huang X. et al. Melatonin reprogramming of gut microbiota improves lipid dysmetabolism in high-fat diet-fed mice. J Pineal Res. 2018;65(4):e12524. doi:10.1111/jpi.12524.
- Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, Zur M, Regev-Lehavi D, Ben-Zeev Brik R, Federici S. et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174(6):1406–1423.e1416. doi:10.1016/j.cell.2018.08.047.
- Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, Kotler E, Zur M, Regev-Lehavi D, Brik RB. et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174(6):1388–1405.e1321. doi:10.1016/j.cell.2018.08.041.
- Kasahara K, Kerby RL, Zhang Q, Pradhan M, Mehrabian M, Lusis AJ, Bergström G, Bäckhed F, Rey FE. Gut bacterial metabolism contributes to host global purine homeostasis. Cell Host & Microbe. 2023;31(6):1038–1053.e10. doi:10.1016/j.chom.2023.05.011.
- Hosomi K, Saito M, Park J, Murakami H, Shibata N, Ando M, Nagatake T, Konishi K, Ohno H, Tanisawa K. et al. Oral administration of blautia wexlerae ameliorates obesity and type 2 diabetes via metabolic remodeling of the gut microbiota. Nat Commun. 2022;13(1):4477. doi:10.1038/s41467-022-32015-7.
- Zhao Z, Ning J, Bao XQ, Shang M, Ma J, Li G, Zhang D. Fecal microbiota transplantation protects rotenone-induced Parkinson’s disease mice via suppressing inflammation mediated by the lipopolysaccharide-TLR4 signaling pathway through the microbiota-gut-brain axis. Microbiome. 2021;9(1):226. doi:10.1186/s40168-021-01107-9.
- Reiter RJ, Mayo JC, Tan DX, Sainz RM, Alatorre-Jimenez M, Qin L. Melatonin as an antioxidant: under promises but over delivers. J Pineal Res. 2016;61(3):253–278. doi:10.1111/jpi.12360.
- Ferreira ZS, Fernandes PA, Duma D, Assreuy J, Avellar MC, Markus RP. Corticosterone modulates noradrenaline-induced melatonin synthesis through inhibition of nuclear factor kappa B. J Pineal Res. 2005;38(3):182–188. doi:10.1111/j.1600-079X.2004.00191.x.
- de Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: mechanistic insights. Gut. 2022;71(5):1020–1032. doi:10.1136/gutjnl-2021-326789.
- Lung TWF, Charytonowicz D, Beaumont KG, Shah SS, Sridhar SH, Gorrie CL, Mu A, Hofstaedter CE, Varisco D, McConville TH. et al. Klebsiella pneumoniae induces host metabolic stress that promotes tolerance to pulmonary infection. Cell Metab. 2022;34(5):761–774.e769. doi:10.1016/j.cmet.2022.03.009.
- Kim G, Yoon Y, Park JH, Park JW, Noh MG, Kim H, Park C, Kwon H, Park JH, Kim Y. et al. Bifidobacterial carbohydrate/nucleoside metabolism enhances oxidative phosphorylation in white adipose tissue to protect against diet-induced obesity. Microbiome. 2022;10(1):188. doi:10.1186/s40168-022-01374-0.
- Pruss KM, Sonnenburg JL. C. difficile exploits a host metabolite produced during toxin-mediated disease. Nature. 2021;593(7858):261–265. doi:10.1038/s41586-021-03502-6.
- Song L, He M, Sun Q, Wang Y, Zhang J, Fang Y, Liu S, Duan L. Roseburia hominis increases intestinal melatonin level by activating p-CREB-AANAT pathway. Nutrients. 2021;14(1). doi:10.3390/nu14010117.
- Tucey TM, Verma J, Harrison PF, Snelgrove SL, Lo TL, Scherer AK, Barugahare AA, Powell DR, Wheeler RT, Hickey MJ. et al. Glucose homeostasis is important for immune cell viability during Candida challenge and host survival of systemic fungal infection. Cell Metab. 2018;27(5):988–1006.e7. doi:10.1016/j.cmet.2018.03.019.
- Song JW, Lam SM, Fan X, Cao WJ, Wang SY, Tian H, Chua GH, Zhang C, Meng FP, Xu Z. et al. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 2020;32(2):188–202.e5. doi:10.1016/j.cmet.2020.06.016.
- McCarville JL, Chen GY, Cuevas VD, Troha K, Ayres JS. Microbiota metabolites in health and disease. Annu Rev Immunol. 2020;38:147–170. doi:10.1146/annurev-immunol-071219-125715.
- Fan L, Xia Y, Wang Y, Han D, Liu Y, Li J, Fu J, Wang L, Gan Z, Liu B. et al. Gut microbiota bridges dietary nutrients and host immunity. Sci China Life Sci. 2023;66(11):2466–2514. doi:10.1007/s11427-023-2346-1.
- Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L. et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23(1):107–113. doi:10.1038/nm.4236.
- Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, Falony G, Raes J, Maiter D, Delzenne NM. et al. Supplementation with akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019;25(7):1096–1103. doi:10.1038/s41591-019-0495-2.
- Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, Kayser BD, Levenez F, Chilloux J, Hoyles L. et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65(3):426–436. doi:10.1136/gutjnl-2014-308778.
- Horvat S, Mahnic A, Breskvar M, Dzeroski S, Rupnik M. Evaluating the effect of Clostridium difficile conditioned medium on fecal microbiota community structure. Sci Rep. 2017;7(1):16448. doi:10.1038/s41598-017-15434-1.
- Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, Van Treuren W, Pruss K, Stabler SR, Lugo K. et al. A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host & Microbe. 2018;24(2):296–307.e7. doi:10.1016/j.chom.2018.07.002.
- Virtue AT, McCright SJ, Wright JM, Jimenez MT, Mowel WK, Kotzin JJ, Joannas L, Basavappa MG, Spencer SP, Clark ML. et al. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRnas. Sci Transl Med. 2019;11(496):11. doi:10.1126/scitranslmed.aav1892.
- Tan J, Ni D, Taitz J, Pinget GV, Read M, Senior A, Wali JA, Elnour R, Shanahan E, Wu H. et al. Dietary protein increases T-cell-independent sIgA production through changes in gut microbiota-derived extracellular vesicles. Nat Commun. 2022;13(1):4336. doi:10.1038/s41467-022-31761-y.
- Luo Z, Ji Y, Gao H, Gomes Dos Reis FC, Bandyopadhyay G, Jin Z, Ly C, Chang YJ, Zhang D, Kumar D. et al. Crig+ macrophages prevent gut microbial DNA–containing extracellular vesicle–induced tissue inflammation and insulin resistance. Gastroenterology. 2021;160(3):863–874. doi:10.1053/j.gastro.2020.10.042.
- Liu Y, Jia Y, Yang K, Tong Z, Shi J, Li R, Xiao X, Ren W, Hardeland R, Reiter RJ. et al. Melatonin overcomes MCR-mediated colistin resistance in Gram-negative pathogens. Theranostics. 2020;10(23):10697–10711. doi:10.7150/thno.45951.
- Karamitri A, Jockers R. Melatonin in type 2 diabetes mellitus and obesity. Nat Rev Endocrinol. 2019;15(2):105–125. doi:10.1038/s41574-018-0130-1.
- Cecon E, Izabelle C, Poder SL, Real F, Zhu A, Tu L, Ghigna MR, Klonjkowski B, Bomsel M, Jockers R. et al. Therapeutic potential of melatonin and melatonergic drugs on K18-hACE2 mice infected with SARS-CoV-2. J Pineal Res. 2022;72(1):e12772. doi:10.1111/jpi.12772.
- Ren W, Liu G, Chen S, Yin J, Wang J, Tan B, Wu G, Bazer FW, Peng Y, Li T. et al. Melatonin signaling in T cells: functions and applications. J Pineal Res. 2017;62(3):62. doi:10.1111/jpi.12394.
- Gomes PRL, Vilas-Boas EA, Leite EA, Munhoz AC, Lucena CF, Amaral FGD, Carpinelli AR, Cipolla-Neto J. Melatonin regulates maternal pancreatic remodeling and B-cell function during pregnancy and lactation. J Pineal Res. 2021;71(1):e12717. doi:10.1111/jpi.12717.
- Xia Y, Chen S, Zeng S, Zhao Y, Zhu C, Deng B, Zhu G, Yin Y, Wang W, Hardeland R. et al. Melatonin in macrophage biology: Current understanding and future perspectives. J Pineal Res. 2019;66(2):e12547. doi:10.1111/jpi.12547.
- Xia Y, Zhang Q, Ye Y, Wu X, He F, Peng Y, Yin Y, Ren W. Melatonergic signalling instructs transcriptional inhibition of IFNGR2 to lessen interleukin-1β-dependent inflammation. Clin Transl Med. 2022;12:e716. doi:10.1002/ctm2.716.
- Álvarez-Sánchez N, Cruz-Chamorro I, López-González A, Utrilla JC, Fernández-Santos JM, Martínez-López A, Lardone PJ, Guerrero JM, Carrillo-Vico A. Melatonin controls experimental autoimmune encephalomyelitis by altering the T effector/regulatory balance. Brain Behav Immun. 2015;50:101–114. doi:10.1016/j.bbi.2015.06.021.
- He F, Liu Y, Li P, Wu X, Xia Y, Zhang D, Li N, Peng Y, Zhu G, Hardeland R. et al. Melatonin inhibits Gram-negative pathogens by targeting citrate synthase. Sci China Life Sci. 2022;65(7):1430–1444. doi:10.1007/s11427-021-2032-9.
- Zhai X, Wang N, Jiao H, Zhang J, Li C, Ren W, Reiter RJ, Su S. Melatonin and other indoles show antiviral activities against swine coronaviruses in vitro at pharmacological concentrations. J Pineal Res. 2021;71(2):e12754. doi:10.1111/jpi.12754.
- Xin Z, Jiang S, Jiang P, Yan X, Fan C, Di S, Wu G, Yang Y, Reiter RJ, Ji G. Melatonin as a treatment for gastrointestinal cancer: a review. J Pineal Res. 2015;58:375–387. doi:10.1111/jpi.12227.
- Liao Y, Fan L, Bin P, Zhu C, Chen Q, Cai Y, Duan J, Cai Q, Han W, Ding S. et al. GABA signaling enforces intestinal germinal center B cell differentiation. Proc Natl Acad Sci. 2022;119(44):e2215921119. doi:10.1073/pnas.2215921119.
- Liu B, Yu D, Sun J, Wu X, Xin Z, Deng B, Fan L, Fu J, Ge L, Ren W. Characterizing the influence of gut microbiota on host tryptophan metabolism with germ-free pigs. Animal Nutr. 2022;11:190–200. doi:10.1016/j.aninu.2022.07.005.
