ABSTRACT
Inulin, an increasingly studied dietary fiber, alters intestinal microbiota. The aim of this study was to assess whether inulin decreases intestinal colonization by multidrug resistant E. coli and to investigate its potential mechanisms of action. Mice with amoxicillin-induced intestinal dysbiosis mice were inoculated with extended spectrum beta-lactamase producing E. coli (ESBL-E. coli). The combination of inulin and pantoprazole (IP) significantly reduced ESBL-E. coli fecal titers, whereas pantoprazole alone did not and inulin had a delayed and limited effect. Fecal microbiome was assessed using shotgun metagenomic sequencing and qPCR. The efficacy of IP was predicted by increased abundance of 74 taxa, including two species of Adlercreutzia. Preventive treatments with A. caecimuris or A. muris also reduced ESBL-E. coli fecal titers. Fecal microbiota of mice effectively treated by IP was enriched in genes involved in inulin catabolism, production of propionate and expression of beta-lactamases. They also had increased beta-lactamase activity and decreased amoxicillin concentration. These results suggest that IP act through production of propionate and degradation of amoxicillin by the microbiota. The combination of pantoprazole and inulin is a potential treatment of intestinal colonization by multidrug-resistant E. coli. The ability of prebiotics to promote propionate and/or beta-lactamase producing bacteria may be used as a screening tool to identify potential treatments of intestinal colonization by multidrug resistant Enterobacterales.
1. Introduction
The gut is the main reservoir of multidrug resistant Enterobacterales (MDRE). Intestinal colonization by MDRE promotes various infections including urinary, gastrointestinal and bloodstream infections, and contaminates the environment and/or healthy or diseased subjects. Different non-antibiotic approaches such as probiotics and fecal microbiota transplantation have been assessed to treat or prevent MDRE intestinal colonization, but treatments that are both effective and easy to administer remain to be identified.Citation1 Although mechanisms of microbiota resistance to intestinal colonization by MDRE after exposure to antibiotics are incompletely elucidated, they include competition for nutrients and inhibition of MDRE by microbiota-produced short chain fatty acids (SCFAs) and more hypothetically isoflavonoids.Citation2–5 Specifically in the case of exposure to a beta-lactam agent, antibiotic degradation in the colonic lumen by beta-lactamase producing microbiota may limit the antibiotic-induced dysbiosis and confer resistance to colonization by MDRE.Citation6–9 Prebiotics are nondigestible carbohydrates that modulate the composition of gut microbiota. Inulin, commercially available as a prebiotic, originates from plant roots.Citation10 Its structure consists of 10–60 units of β-D-fructosyl subgroups linked together by (2→1) glycosidic bonds that are not digested by gut enzymes. Inulin is selectively fermented by certain intestinal bacteria, and induces the production of SCFAs in the colon, including butyrate, acetate and propionate.Citation2,Citation11,Citation12 Indeed, in humans, inulin promotes intestinal growth of SCFA producers, including genera of various families such as Bifidobacteriaceae (Bifidobacterium), Lachnospiraceae (Anaerostipes), Oscillospiraceae (Faecalibacterium) and Lactobacillaceae (Lactobacillus).Citation13 Furthermore, in mice inulin promotes the intestinal growth of Lachnospiraceae (Blautia, Roseburia), Muribaculaceae (Duncaniella muris), Oscillospiraceae (Ruminococcus) and Akkermansiaceae (Akkermansia) that are also associated with resistance to colonization by MDRE in antibiotic dysbiosed mice.Citation14–21 Hence, we made the hypothesis that, by altering intestinal microbiota and promoting SCFA producers, inulin would limit the level and/or prevalence of intestinal colonization by MDRE.
The aim of this study was to assess whether inulin prevents intestinal colonization by an extended spectrum beta-lactamase producing E. coli (ESBL-E. coli) in mice with amoxicillin-induced intestinal dysbiosis. We also investigated potential mechanisms by which this treatment could reduce MDRE colonization, including the increased abundance of inulin catabolizing and SCFA producing bacteria, equol production and decreased fecal concentration of amoxicillin by beta-lactamase producing bacteria, resulting in the limitation of antibiotic-induced dysbiosis. In our model, pantoprazole was used to decrease gastric acidity and promote the colonization by ESBL-E. coli.Citation22,Citation23 We found that the combination of inulin and pantoprazole decreased the level of ESBL-E. coli colonization, and identified various predictors of treatment efficacy.
2. Methods
2.1. Bacterial strains and media
A clinical isolate of phylogroup B2, serotype O25, ESBL-producing -E. coli was collected from University Hospital of Nantes (GenBank accession number: JAOTNQ000000000). In addition to ESBL production mediated by CTX-M-27, it was also resistant to carbapenem antibiotics through an OXA-181 carbapenemase. ESBL-E. coli enumeration was realized on ChromIDTM ESBL agar plates (Biomerieux). Adlercreutzia strains were grown under anaerobic conditions (N2-CO2, 80:20) on supplemented BHI medium (Oxoid) with glucose 0.4% (Braun), cysteine 0.05% (Sigma-Aldrich), and yeast extract 0.04% (Biokar).Citation24
2.2. Murine model of ESBL-E. coli intestinal colonization
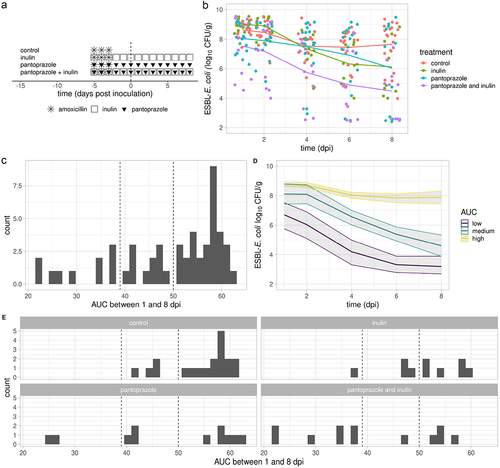
All experiments were approved by the Animal Experiment Committee of Pays de la Loire (France, authorization number APAFIS#18120) and complied to ARRIVE Guidelines. Six weeks old male Swiss mice (Janvier Labs, Saint-Berthevin, France) were housed individually to avoid inter-individual contamination, in pathogen free conditions with free access to food and water. Intestinal microbiota was first altered by amoxicillin administration in drinking water (0.5 g·L−1) for 3 days to make mice susceptible to ESBL-E. coli colonization (). Inulin sourced from chicory roots was purchased from Bulk Powders. A 0.2% (w/w) solution was prepared by dissolving inulin powder in distilled water before autoclaving, and administered in drinking water from the first day of amoxicillin. In some mice, pantoprazole was added to drinking water (0.1 g·L−1) on the first day of amoxicillin, because it is known to suppress the gastric acid production and to promote intestinal colonization by multidrug resistant bacteria.Citation22 Two days after amoxicillin discontinuation, ESBL-E. coli was intragastrically inoculated (10Citation6 CFU per mouse). Mice were monitored for 8 days and feces were serially collected. Mice were randomly divided into four groups () : control (n = 19); inulin (n = 10); pantoprazole (n = 11); inulin and pantoprazole (IP, n = 12). About 50 mg of feces were suspended in 1 ml of sterile water and shaken at 20 Hz (Mixer Mill MM 400, RETSCH’s) for 5 min. ESBL-E. coli fecal titers were assessed after incubation at 37°C for 24 hr in aerobic conditions.
Figure 1. Effect of inulin, pantoprazole and their combination on ESBL-E. coli fecal titers.
2.3. Beta-Lactamase detection assay and amoxicillin fecal concentrations
Feces sampled 2 days before ESBL-E. coli inoculation were resuspended in 1 ml of phosphate-buffered saline (PBS). Samples were left for 5 min to allow particulate matter to sediment. Next, 180 µl of the suspension were incubated 30 min at 37°C with 20 µl of Nitrocefin 0.5 mg/ml (Merck). Then the β-Lactamase activity was determined in the supernatant by absorption at 492 nm. Amoxicillin fecal concentrations were assessed as follows. Feces sampled 2 days before ESBL-E.coli inoculation were weighed, resuspended and homogenized in water (10 mg/100 μL) by sonication. The material obtained was centrifuged (5 min 13,000 g, + 4 ° C). The supernatant was mixed with acetonitrile solution containing the internal standard (13C6 amoxicillin). After centrifugation of the mixture (5 min 13,000 g, +4°C), the supernatant was diluted in formic acid 0.1% v/v (1/2 v/v) and injected into the chromatographic system. The system consisted of a Kinetex® 2.6 µm C18 50 mm x 2.1 mm column (Phenomenex, Le Pecq, France) in a thermostatically controlled oven at 40°C, mobile phases with a binary gradient [(acetonitrile/formic acid 0.1% v/v) and (ultrapure water/formic acid 0.1% v/v)] at a flow rate of 0.5 mL/min and a tandem mass spectrometry monitoring (3200 QTRAP® Sciex, Villebon-sur-Yvette, France).
2.4. Microbiome analysis using quantitative PCR
DNA from fecal samples (30 mg) was extracted with Nucleospin DNA stool kit (Macherey Nagel) and eluted in 30 µl of buffer. We used previously published primers (Supplementary material, table S6). We also designed primers for Bacteroides acidifaciens, Duncaniella muris and Adlercreutzia caecimuris. Primers for the specific amplification of Bacteroides acidifaciens 5’-CGATGAAGACGGAAGAAGTGG (Baci3) and 5’-TTCAAGTTCATAAAGCTCATCATTC (Baci4) were designed as follows: B. acidifaciens genomes were acquired from NCBI public database. Coding sequences (CDSs) from these genomes were clustered together by CD-HIT v4.8.1 at a 95% nucleotide identity threshold in global alignment.Citation25 Genes that were present in all genomes were taken for further analysis. CDSs of other species (off-targets) were downloaded from the NCBI. Conserved genes from B. acidifaciens were clustered with the genes from most similar species at the 80% nucleotide identity threshold. Sequences that clustered together with off-targets were discarded. The target gene was selected and searched for dissimilarity by BLAST in NCBI. Primers were designed on the NCBI web server using Primer-BLAST and checked for homo- and heteroduplexes using the OligoAnalyzer® tool from Integrated DNA Technologies, Inc.Citation26 Those for Duncaniella muris 5’-TCACCATCCGTGAGATGCCTCC (Dunca-f), 5’- ATAGAGGAAAGCCGCCCAGCAG (Dunca-r) and Adlercreutzia caecimuris 5’-AGTCACGCACCCCCGTATTCTC (ACA-f), 5’-CGCGCCATTCGATGATGCTTCC (ACA-r) were designed by comparing the genome of D. muris and A. caecimuris with the genome of 25 bacteria specially selected for the identification of unique sequences. We selected bacteria affiliated or not with the same taxonomic genus in order to achieve the widest possible analysis. All bacterial genomes were acquired from NCBI public database with the Entrez Direct (EDirect) tools. These genomes were used to develop a BLAST database and then to compare genomes using «blastn» command lines in the BASH script available on https://github.com/ncbi/blast_plus_docs. The unique sequences were retrieved and quality control was performed using the NCBI BLASTn and UniProt BLAST tools. The primers have been designed by the Eurofins Genomics webserver «PCR Primer Design Tool» (https://eurofinsgenomics.eu/en/ecom/tools/pcr-primer-design/). The selection criteria for the primers were as follows: a melting temperature close to 60°C, a percentage of GC greater than 50% and an amplicon length between 300 and 500 bps. Their specificity were controlled by PCR amplification from fecal DNA and sequencing of the amplicon.
The PCR reactions were performed in a total volume of 20 µl. All taxa were detected by using the PowerUp SYBR Green Master Mix (Life Technologies), with 10 pmol of each of the forward and reverse primers for each reaction. Most of the PCR reaction conditions were 94°C for 3 min, then 40 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec. For the PCR of Bacteroides species, EPC, D. muris and A. caecimuris the conditions were the following : 94°C for 3 min, then 40 cycles of 94°C for 30 sec, 59°C for 1 min. The calibrations were done with each PCR fragment amplified from the respective strains and whose concentration was adjusted to 5.106 molecules/µl. The specificity of each primer pair was verified by controlling that only one band of the expected size was obtained on a 2% agarose gel after amplification from the fecal DNA and that its sequence was as expected. The titers of each bacteria was inferred from the shift of the threshold cycle (CT), obtained by amplifying target from the fecal DNA in comparison to that of reference DNA. Corrections were made when the target was the 16S RNA gene since it is differently repeated in the bacterial genomes.
2.5. Metagenomic analyses
Fresh stools samples were immediately frozen and stored at − 80°C until DNA extraction. The DNA was then extracted using the PowerSoil Pro HTP (Qiagen Inc., Venlo, The Netherlands) following the manufacturer’s protocol. Sequencing was then performed using NovaSeq S1 150PE platform at the University of Minnesota Genomic Center. Raw sequences were filtered and trimmed using FastQC (v. 0.11.9) and Trimmomatic (v. 0.36).Citation27 Metagenomic taxonomic classification was performed using Kraken2 (v. 2.1.2) and Bracken (v. 2.5.0) against the mouse specific MGBC database (release 2.0).Citation28–30 For antibiotic resistance gene (ARG) quantification, ARG-OAP v.2.0 pipeline was used.Citation31 Subsampled FASTQ files were also processed with ARG-OAP v.2.0 to obtain the annotation of ARG profiles. ARG-OAP v.2.0 provides model-based identification of assembled sequences using SARGfam, a high-quality profile Hidden Markov Model containing profiles of ARG subtypes and including cell number quantification by using the average coverage of essential single-copy marker genes. ARG abundances were normalized by cell number. Each reference sequence was tagged with its functional gene annotation (ARG subtype) and membership within a class of antibiotics targeted by the gene (ARG type). Sequencing data are available in the NCBI Sequence Read Archive and publicly accessible under BioProject ID PRJNA1052115.
2.6. Statistical analyses
Median are reported with 1st and 3rd quartiles, and means with SD. Treatment efficacy at different time points was assessed through comparison of ESBL-E. coli fecal titers using linear mixed effect models. Area under the curve of ESBL-E. coli fecal titers was calculated by the trapezoidal method between 1 and 8 dpi. Diversity analyses were performed using vegan R package (version 2.5–7). Taxa which were not present in at least 20% of the samples were removed. Beta diversity was assessed from Bray-Curtis distances that were compared with permutational multivariate analysis of variance (PERMANOVA). Differences in fecal microbiome composition among samples can be visualized by means of the Principal Coordinates Analysis (PCoA), such that each point represents a single fecal sample. Similar samples are located relatively close to each other, and clusters of distinct microbiome compositions can be appreciated. We defined potential predictors of treatment efficacy as features that were significantly different between effectively treated mice and control or ineffectively treated mice (FDR corrected Wilcoxon test p value ≤ .05 with absolute value of generalized fold change > 0.5 log10). All statistical analyses were performed with R 3.6.3, R Foundation for Statistical Computing, Vienna, Austria.
3. Results
3.1. The combination of inulin and pantoprazole limited the intestinal dysbiosis
Intestinal dysbiosis is considered to be necessary to achieve a sustained colonization by ESBL-E. coli in mice. Mice were exposed to amoxicillin during 3 days. After a 2-day wash-out they were intragastrically inoculated with ESBL-E. coli (). To show that amoxicillin induced intestinal dysbiosis, we used qPCR to assess the effect of amoxicillin on fecal microbiota of control mice, by comparing feces sampled 5 days before starting amoxicillin, that is, at −5 days post-inoculation (dpi, naive feces) and before ESBL-E. coli inoculation (0 dpi). Amoxicillin decreased significantly DNA fecal concentration and titers of Bacteroidota, Adlercreutzia caecimuris, Bacteroides acidifaciens, B. ovatus, Duncaniella muris, Muribaculum sp. and Equol producing Coriobacteriia (EPC), and increased Enterococcus (Extended data, Figure S1).
Then, we assessed the impact of treatments on fecal microbiome by comparing feces sampled just before ESBL-E. coli inoculation (0 dpi) in control and treated mice. The inulin-pantoprazole treatment increased DNA fecal concentration and abundance of 8 taxa (Bacteroidota, Bacteroides, B. acidifaciens, A. caecimuris, EPC, B. ovatus, B. thetaiotaomicron and E. coli), of which 6 had been reduced by amoxicillin (Extended data, Figure S1). Pantoprazole alone increased DNA fecal concentration and abundance of 4 taxa that were decreased by amoxicillin (Bacteroidota, Muribaculum sp., Duncaniella muris and EPC). Inulin alone induced no significant changes in DNA fecal concentration or selected taxa, including Lactobacillus sp. and Bifidobacterium sp. previously reported to be increased by inulin (data not shown). Hence, qPCR analyses of fecal microbiome showed that the combination of inulin and pantoprazole, and to a lesser extent pantoprazole alone, limited the amoxicillin-induced intestinal dysbiosis.
3.2. The combination of inulin and pantoprazole decreased fecal titers of ESBL-E. coli
The efficacy of different treatments was assessed by comparing mean ESBL-E. coli fecal titers in treated and control mice using a multivariate model between 1 and 8 dpi (Extended data, Table S1). In the control group, ESBL-E. coli titers slightly decreased with time, by 0.2 ± 0.1 log10 CFU/g/day. In inulin-treated mice, ESBL-E. coli titers tended to be higher than control (9.0 ± 0.4 vs 8.6 ± 0.5 log10 CFU/g, Wilcoxon test P-value, .06) at 1 dpi, but they subsequently decreased by 0.3 ± 0.1 log10 CFU/g/day, resulting in lower titers at 8 dpi (6.1 ± 1.9 vs 7.7 ± 1.6 log10 CFU/g, Wilcoxon test P-value, .03, ). Pantoprazole alone had no significant activity on ESBL-E. coli titers. The combination of inulin and pantoprazole (IP) showed a better efficacy than single treatments. Indeed, it significantly decreased ESBL-E. coli titers by 0.7 ± 0.3 log10 CFU/g and the effect gradually increased over time by 0.3 ± 0.1 log10 CFU/g each day (, Table S2). Finally, at 8 dpi, mean titers were lower in IP-treated than in control mice (4.5 ± 1.8 vs 7.7 ± 1.6 log10 CFU/g, Wilcoxon test P-value, <.001).
Inspection of ESBL-E. coli titers of each mice showed that treatments were highly effective in some mice, and not effective in other mice. To support this observation, we computed for each mouse the AUC of ESBL-E. coli titers between 1 and 8 dpi (AUCESBL-E. coli). AUCESBL-E. coli ranged between 22 and 64 log10 CFU·day·g−1 (). AUCESBL-E. coli were grouped into three classes in order to identify treated mice with lower ESBL-E. coli titers than any control mice. From the distribution of AUCESBL-E. coli in various groups of treatment, we defined low, medium and high levels of ESBL-E. coli colonization as AUCESBL-E. coli ≤39, between 39 and 50 or ≥ 50 log10 CFU·day·g−1, respectively (). The proportion of mice with low AUCESBL-E. coli in control and IP treated mice was 0 among 19 (0%) and 7 among 12 (58%), respectively (P-value, < .001). The proportion of low AUCESBL-E. coli in mice treated with IP (58%) was higher than in mice treated with either pantoprazole or inulin (14%; chiCitation2 P-value, .02). Hence, this classification of AUCESBL-E. coli identified mice effectively treated by any treatment, and confirmed the better efficacy of IP over pantoprazole or inulin alone.
In the remainder of this study, we sought to identify factors predictive of the effectiveness of the combination of inulin and pantoprazole, by comparing mice effectively treated by IP (effective treatment defined by low AUCESBL-E. coli) and control mice or mice ineffectively treated by IP (medium or high AUCESBL-E. coli).
3.3. The microbiome architecture predicted the treatment efficacy
To identify taxa predictive of the efficacy of IP, we analyzed fecal microbiome diversity and composition before ESBL-E. coli inoculation (i.e. at 0 dpi). Fecal microbiome was assessed using shotgun metagenomic sequencing in control mice (n = 7, including 6 high and 1 medium AUCESBL-E. coli) and mice treated with IP (n = 11, including 4 high, 1 medium and 6 high AUCESBL-E. coli). We grouped together control and ineffectively treated mice (high or medium AUCESBL-E. coli, n = 12) and compared them to effectively treated mice (low AUCESBL-E. coli, n = 6).
Alpha-diversity was higher in effectively treated mice (Shannon index, P-value, .004; observed species, P-value, .02, ). Bray-Curtis dissimilarities readily separated the fecal microbiome of effectively treated mice and control or ineffectively treated mice (PERMANOVA, P-value, .001), and the first two principal coordinates discriminated the two groups of mice (). Among 195 species, 74 were differentially abundant between the two groups, all being increased in effectively treated mice (Extended data, Table S2). More specifically, mice effectively treated with IP had higher titers of 28 species of the Muribaculaceae family (including Duncaniella muris and 2 other Duncaniella sp., Muribaculum intestinale and another Muribaculum sp., Paramuribaculum intestinale and 2 other Paramuribaculum species), 16 species of the Lachnospiraceae family, 6 species of order Enterobacterales (including E. coli, Citrobacter A rodentium, Klebsiella pneumoniae, K. oxytoca and K. variicola), 6 species of genus Alistipes (family Rikenellaceae), 3 species of the Coriobacteriia class and Eggerthellaceae family (Adlercreutzia caecimuris, Adlercreutzia_sp004793465 and a Parvibacter sp.), and 2 species of the Bacteroidaceae family (including one Bacteroides).
Figure 2. Taxonomic diversity and architecture of fecal microbiome in mice effectively treated by inulin and pantoprazole in comparison with control or ineffectively treated mice.
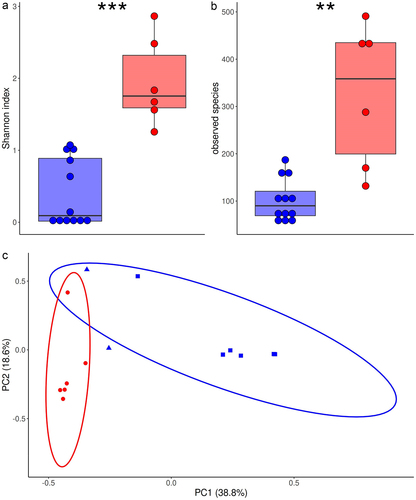
To support these results based on relative abundances, we assessed fecal titers of selected taxa with qPCR. In comparison with control or ineffectively treated mice, mice effectively treated with IP had significantly higher DNA fecal concentration (median, 99 (71–147) vs 27 (22–45) µg/g, p = .002 and higher titers of Muribaculum sp., Duncaniella muris, E. coli and A. caecimuris, thus confirming results of metagenomic sequencing (Extended data, Figure S2). EPC, Bacteroides and Akkermansia had also higher titers, and Enterococcus lower titers in mice effectively treated with IP.
3.4. Treatment with adlercreutzia decreased ESBL-E. coli colonization
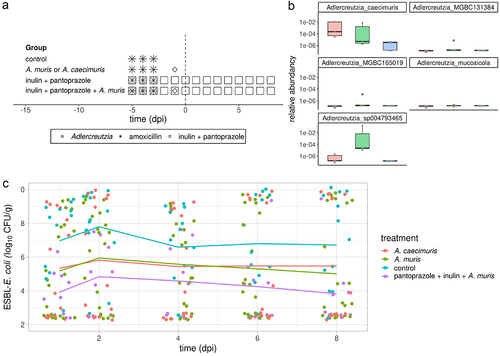
Quantitative PCR and metagenomic analyses showed that higher titers of EPC, Adlercreutzia caecimuris and Adlercreutzia_sp004793465 were predictive of efficacy of IP. Hence, we assessed whether treating mice with Adlercreutzia caecimuris or any equol-producing Adlercreutzia would decrease the level of ESBL-E. coli colonization. To isolate a murine isolate of equol producing Adlercreutzia, we selected a mouse with a high titer of EPC at 0 dpi and subsequently colonized at low level by ESBL-E. coli. Fecal samples were cultivated on a medium designed to isolate Adlercreutzia containing colimycin 20 µg/ml. Several colonies were tested by PCR with the EPC primers and a positive colony was further characterized. Its 16S RNA gene sequence had 99% identity on 900bp with that of Adlercreutzia muris strain SP-7 (DSM 29,508), and genome sequencing identified the isolate as A. muris (NCBI accession number JAOTNR000000000). We also purchased the A. caecimuris DSM 21,839. In a new set of experiments, mice were treated with 7 log10 CFU of either A. caecimuris DSM 21,839 or A. muris. Shotgun metagenomic sequencing showed higher A. caecimuris relative abundancy in mice treated with A. caecimuris than in control mice (). The five species of Adlercreutzia identified by shotgun metagenomic analysis did not include A. muris, but Adlercreutzia_sp004793465 was detected in all mice treated with A. muris and in none of the control mice, suggesting that Adlercreutzia_sp004793465 belongs to the A. muris species (). In comparison with control mice, preventive treatments with A. caecimuris and A. muris decreased significantly ESBL-E. coli titers by 1.6 ± 0.4 and 1.5 ± 0.4 log10 CFU/g, respectively, without significant variation of effect over time (; Extended data, table S3). The combination of inulin, pantoprazole and A. muris was more effective, decreasing ESBL-E. coli titers by 2.7 ± 0.5 log10 CFU/g. Mice treated with the combination of A. muris and IP had lower ESBL-E. coli titers than those treated by A. muris alone (difference, −1.2 ± 0.5 log10 CFU/g, p = .035) or by IP alone (difference, −3.4 ± 0.6 log10 CFU/g, p < .0001). Hence, these results support the hypothesis that Adlercreutzia sp. is involved in the activity of IP treatment.
Figure 3. Preventive treatment with adlercreutzia caecimuris or adlercreutzia muris on ESBL-E. coli intestinal colonization.
3.5. Higher abundance of genes involved in inulin catabolism and propionate production predicted the treatment efficacy
Then we searched for associations between efficacy of IP and metabolic pathways of fecal microbiome at 0 dpi. Metabolic pathways were predicted using HUMAnN3 and the MetaCyc Metabolic Pathways database.Citation32,Citation33 Alpha-diversity of metabolic pathways was higher in effectively treated mice (Shannon index, P-value, .067; observed species, P-value, .003, ). The first two principal coordinates discriminated effectively treated mice and control or ineffectively treated mice (), and PERMANOVA showed significant difference in their overall architecture (P-value, .001). Among 301 metabolic pathways, 85 were differentially abundant between effectively treated mice and control or ineffectively treated mice (Extended data, table S4). Of note, 9 of these pathways were involved in synthesis of propionate, all being higher in effectively treated mice (Extended data, Figure S3). None was directly associated with inulin catabolism.
Figure 4. Metabolic pathways of fecal microbiota in mice effectively treated with inulin and pantoprazole vs control or ineffectively treated mice.
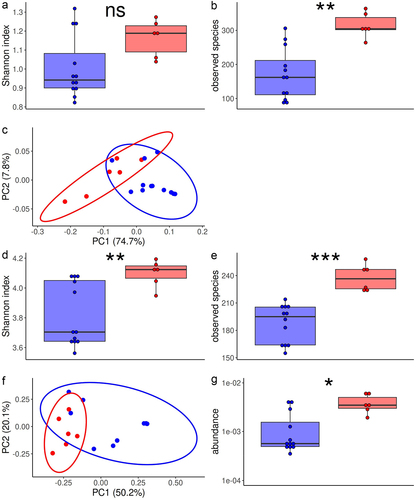
To complete this analysis, we used the CAZy database (www.cazy.org), specifically dedicated to carbohydrate metabolism.Citation34 Alpha-diversity was higher in effectively treated mice (Shannon index, P-value, .007; observed species, P-value, <.001, ). The first two principal coordinates discriminated effectively treated mice and control or ineffectively treated mice (), and PERMANOVA showed significant difference in their overall architecture (P-value, .003). The relative abundance was significantly different for 94 among 225 families or modules of carbohydrate active enzymes, including 72 increased and 22 decreased relative abundances in effectively treated mice in comparison with control or ineffectively treated mice (Extended data, Table S5). Among those, gene abundance of fructan beta-fructosidase, that is involved in catabolism of inulin, was higher in feces of effectively treated mice than in control or ineffectively treated mice ().
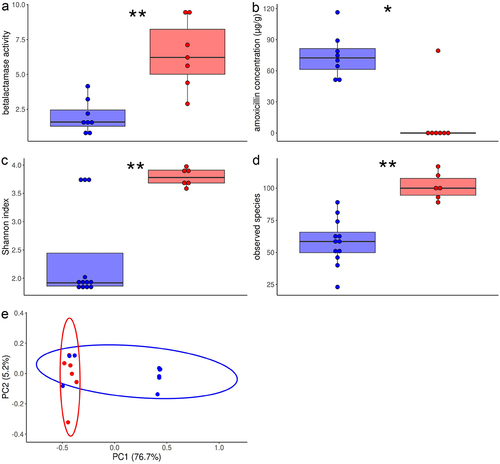
3.6. Higher β-lactamase activity and abundance of β-lactam resistance genes, and lower amoxicillin fecal concentration predicted the treatment efficacy
Since β-lactamase producing bacteria have been reported to decrease amoxicillin fecal concentration and prevent the effect of amoxicillin on the microbiota, we assessed β-lactamase activity and amoxicillin concentration in the feces sampled just after amoxicillin discontinuation (−2 dpi).Citation6 In comparison with control mice, IP increased fecal β-lactamase activity (median (Q1-Q3), 5.0 (3.5–6.9) AU/g vs 1.4 (0.8–1.5) AU/g, p = .003) and decreased amoxicillin concentration (median (Q1-Q3), 0 (0–69) µg/g vs 79 (52–89) µg/g, p = .05). Fecal amoxicillin concentration and β-lactamase activity were significantly correlated (Spearman ρ, −0.73, p = .002). Moreover, mice treated by pantoprazole alone had higher fecal β-lactamase activity (median, 3.3 [3.0–6.2] AU, Wilcoxon test P-value, p = .01) and lower amoxicillin concentration (median, 0 [0–0] µg/g, Wilcoxon test P-value, p = .13) than control mice. Mice effectively treated with IP had higher fecal β-lactamase activity and lower amoxicillin concentration than control or ineffectively treated mice (). Furthermore, 5 Enterobacterales isolates randomly picked from feces sampled 2 days before ESBL-E. coli inoculation from five mice with low level of colonization by ESBL-E. coli were resistant to amoxicillin.
Resistome alpha-diversity was higher in mice effectively treated with IP than in control or ineffectively treated mice (ARG subtypes, Wilcoxon test p-value, .01 for Shannon index, 0.001 for Observed Species, ). Bray-Curtis dissimilarities readily separated the fecal resistome from effectively treated mice and control or ineffectively treated mice (PERMANOVA, p-value = .004, ). We identified 19 ARG subtypes that were differentially abundant between effectively treated mice and control or ineffectively treated mice (Extended data, Figure S4). Among these 19 ARG subtypes, 13 conferred resistance to betalactam antibiotics, 12 of them coding for a SHV beta-lactamase and being higher in effectively treated mice. Most of these beta-lactamase resistance genes code for enzymes that hydrolyze amoxicillin. Furthermore, abundance of beta-lactam ARG was correlated with fecal beta-lactamase activity (Spearman rho, 0.57, P-value, .04).
Figure 5. Fecal β-lactamase activity, amoxicillin concentration and resistome in mice effectively treated by inulin and pantoprazole and in control or ineffectively treated mice.
4. Discussion
Here, we showed that a preventive treatment with a combination of inulin and pantoprazole synergistically decreased ESBL-E. coli intestinal colonization in amoxicillin dysbiosed mice. Inulin without pantoprazole did not limit amoxicillin-induced dysbiosis, as assessed by qPCR, and this result was consistent with the limited and delayed efficacy of inulin on ESBL-E. coli colonization. Unlike inulin alone, pantoprazole and the combination of inulin and pantoprazole limited the dysbiosis induced by amoxicillin. We initially used pantoprazole to decrease gastric acidity and promote high level colonization.Citation22 In our study, pantoprazole-treated mice had no higher ESBL-E. coli titers than control mice, probably because control mice had high ESBL-E. coli titers due to high inoculum, in line with what we recently reported with a different murine model.Citation35 We hypothesize that pantoprazole helped to maintain the richness and diversity of the microbiota exposed to amoxicillin, including beta-lactamase-producing bacteria, which would decrease the intraluminal concentration of amoxicillin and attenuate the effect of amoxicillin on microbiota. Pantoprazole may also decrease the susceptibility of microbiota to antibiotics by stimulating efflux pumps, as reported for tigecycline.Citation36 The impact of proton pump inhibitors (PPI) on intestinal microbiota and digestive colonization by MDRE remain debated.Citation37–39 Some studies found that PPI decreased alpha-diversity while others did not.Citation40 In agreement with our results, a small-size study previously found that PPI use was associated with higher alpha diversity in older inpatients treated with more than two antibiotics, suggesting that PPIs may prevent antibiotic-induced dysbiosis.Citation41 PPI effects on microbiota and intestinal colonization by MDRE in antibiotic treated patients may depend on duration of PPI treatment and delay between PPI and antibiotic treatments, but these points remain to be elucidated.
The interindividual variability of IP efficacy may be viewed as a pitfall for its therapeutical use, but it gave us the opportunity to search predictors of IP efficacy in order to decipher its mode of action. Taxonomic and functional analyses showed that IP efficacy was associated with increased abundances of taxa known to be promoted in inulin-fed mice (in particular, taxa of Muribaculaceae and Lachnospiraceae families) as well as the gene of fructan beta-fructosidase that is involved in inulin metabolism, reinforcing the plausibility of its effect on ESBL-E. coli colonization.Citation13 Indeed, the Muribaculaceae family has been reported to be predominant in murine cecal microbiota, with high level expression of genes involved in carbohydrate metabolism and production of SCFAs, especially propionate.Citation42–44 Microbiota-produced SCFAs prevent digestive colonization by MDRE.Citation2 Consistently, among predictors of IP efficacy, we found several taxa known to produce SCFAs, including Duncaniella muris and other Muribaculaceae, Alistipes and species of the Lachnospiraceae family, and 9 predicted metabolic pathways involved in propionate synthesis.Citation45–47 Hence, the IP combination may decrease ESBL-E. coli titers through the production of SCFAs, especially propionate, from inulin fermentation.
The IP combination may also prevent ESBL-E. coli colonization by increasing intestinal beta-lactamase activity through an increased abundance of SHV beta-lactamases (associated with Enterobacterales as supported by the resistance to amoxicillin observed in Enterobacterales isolated from feces of mice with low-level ESBL-E. coli colonization), thus decreasing intestinal concentrations of amoxicillin and microbiota susceptibility to colonization by ESBL-E. coli. This mechanism of action is supported by previous reports that show that various beta-lactamase producing bacteria reduced beta-lactam induced gut dysbiosis and colonization by multidrug organisms.Citation6–9 However, this mechanism seems not sufficient to explain the activity of IP on ESBL-E. coli colonization, as pantoprazole altered fecal beta-lactamase activity and amoxicillin concentration, without decreasing ESBL-E. coli titers. Of note, we found that IP-induced increase of resistome α-diversity and abundances of 18 ARG was associated with a beneficial effect on ESBL-E. coli colonization. This type of counter-intuitive result had previously been reported in a study that showed that a diet rich in non digestible carbohydrates decreased the abundancy of 86 ARG in obese children, but also increased the abundancy of 10 other ARG.Citation48 Similarly, probiotics may have various effects on gut resistome, although those are as poorly described as for prebiotics. For example, a 11-strain probiotic administered after antibacterial therapy increased resistome α-diversity in murine cecal luminal and in human colonic mucosal microbiota.Citation49 In a context where knowledge of relationships between resistome and health outcomes remains preliminary, these results encourage us not to consider that an increase in the diversity of the resistome or in the abundance of certain ARGs is necessarily detrimental to health. Another mechanism of action may be the competition for nutrients between the IP promoted Enterobacterales, especially Klebsiella sp., and the inoculated ESBL-E. coli, as previously shown with K. michiganensis or K. oxytoca.Citation3,Citation4 In line with these results, we found that high abundancy of three species of Klebsiella sp. (K. pneumoniae, K. oxytoca and K. variicola) was predictive of the efficacy of IP. Finally, equol production may be involved in the activity of IP on ESBL-E. coli colonization. Equol is an isoflavonoid produced from dietary daidzein by several taxa, mainly of the Coriobacteriiia class, including Adlercreutzia equolifaciens.Citation50 Equol inhibits the in vitro biofilm formation by carbapenem resistant E. coli.Citation5 qPCR of the tdr gene showed that an increased titer of EPC predicted IP efficacy, as well as relative abundances of three Coriobacteriia including two Adlercreutzia sp. The tdr gene was present in the genome of the A. muris strain that we successfully used to treat ESBL-E. coli colonization, but not in the A. caecimuris DSM21839 genome. Furthermore, equol production has been described in non Coriobacteriia such as Alistipes and Eubacterium, that we also found to be associated with IP efficacy.Citation51,Citation52 Hence, the efficacy of treatment with A. muris, IP and their combination may be partly mediated by equol production. To summarize our current understanding of its mechanisms of action, the IP combination limited amoxicillin-associated dysbiosis by promoting bacterial species that are able to metabolize inulin and/or to hydrolyze amoxicillin in the colonic lumen through the production of betalactamases. This limited dysbiosis likely prevented MDRE colonization by nutrient competition at least with endogenous Enterobacterales such as Klebsiella as previously reported, but also by the production of SCFA and more hypothetically of equol.
This work presents several limitations. First, we did not assess fecal or cecal concentrations of SCFAs to confirm that propionate concentration is predictive of the level of colonization by ESBL-E. coli. Second, we could not determine taxa involved in the increased abundance of beta-lactamase genes and activity. In future studies, identification of amoxicillin resistant Enterobacterales on specific culture media would help to ascertain their involvement in the efficacy of IP. Third, we did not elucidate why the Enterobacterales titer increased in mice effectively treated with IP. Fourth, the extrapolation of our results in antibiotic-treated patients must be prudent because there are numerous differences in gut structure, intestinal microbiota composition and metabolic pathways between mice and Humans. For example, the Muribaculaceae family, highly involved in IP efficacy in mice, is far less abundant in human than in murine microbiota. However, it is plausible that, as we showed in mice, the efficacy of IP in Humans may show interindividual variability, and that the multidimensional analysis of microbiota (including microbiome architecture, resistome and beta-lactamase activity, fecal concentration of antibiotic and/or SCFA production) may help to predict treatment efficacy.
4.1. Conclusion
Our results showed that inulin had a limited and delayed preventive effect on ESBL-E. coli colonization in a murine model of amoxicillin-induced dysbiosis. By contrast, inulin combined with pantoprazole had a synergistic, early effect on intestinal colonization by ESBL-E. coli. Our results suggest that the therapeutic activity of the combination of inulin and pantoprazole may be mediated by various mechanisms, including the degradation of amoxicillin in the intestinal content through promotion of beta-lactamase-producing taxa, inulin-mediated production of SFCA and the production of equol. These results open new insight for the future use of inulin combined with pantoprazole for the prevention of ESBL-E. coli colonization. Further studies are needed to determine the applicability of this treatment in humans.
Supplemental Material
Download MS Word (4.6 MB)Acknowledgments
We are most grateful to the Genomics Core Facility GenoA, member of Biogenouest and France Genomique and to the Bioinformatics Core Facility BiRD, member of Biogenouest and Institut Français de Bioinformatique (IFB) (ANR-11-INBS-0013) for the use of their resources and their technical support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2024.2347021
Additional information
Funding
References
- Campos-Madueno EI, Moradi M, Eddoubaji Y, Shahi F, Moradi S, Bernasconi OJ, Moser AI, Endimiani A. Intestinal colonization with multidrug-resistant Enterobacterales: screening, epidemiology, clinical impact, and strategies to decolonize carriers. Eur J Clin Microbiol Infect Dis. 2023;42(3):229–16. doi:10.1007/s10096-023-04548-2.
- Sorbara MT, Dubin K, Littmann ER, Moody TU, Fontana E, Seok R, Leiner IM, Taur Y, Peled JU, van den Brink MRM. et al. Inhibiting antibiotic-resistant enterobacteriaceae by microbiota-mediated intracellular acidification. J Exp Med. 2019;216(1):84–98. doi:10.1084/jem.20181639.
- Oliveira RA, Ng KM, Correia MB, Cabral V, Shi H, Sonnenburg JL, Huang KC, Xavier KB. Klebsiella michiganensis transmission enhances resistance to enterobacteriaceae gut invasion by nutrition competition. Nat Microbiol. 2020;5(4):630–641. doi:10.1038/s41564-019-0658-4.
- Osbelt L, Wende M, Almási É, Derksen E, Muthukumarasamy U, Lesker TR, Galvez EJC, Pils MC, Schalk E, Chhatwal P. et al. Klebsiella oxytoca causes colonization resistance against multidrug-resistant K. pneumoniae in the gut via cooperative carbohydrate competition. Cell Host Microbe. 2021;29(11):1663–1679.e7. doi:10.1016/j.chom.2021.09.003.
- Kim H-R, Eom Y-B. Synergistic activity of Equol and meropenem against Carbapenem-Resistant Escherichia coli. Antibiotics (Basel). 2021;10(2):161. doi:10.3390/antibiotics10020161.
- Gjonbalaj M, Keith JW, Do MH, Hohl TM, Pamer EG, Becattini S. Antibiotic degradation by commensal microbes shields pathogens. Infect Immun. 2020;88(4):e00012–20. doi:10.1128/IAI.00012-20.
- Léonard F, Andremont A, Leclerq B, Labia R, Tancrède C. Use of -lactamase-producing anaerobes to prevent ceftriaxone from degrading intestinal resistance to colonization. J Infect Dis. 1989;160(2):274–280. doi:10.1093/infdis/160.2.274.
- Stiefel U, Nerandzic MM, Pultz MJ, Donskey CJ. Gastrointestinal colonization with a cephalosporinase-producing bacteroides species preserves colonization resistance against vancomycin-resistant enterococcus and clostridium difficile in cephalosporin-treated mice. Antimicrob Agents Chemother. 2014;58(8):4535–4542. doi:10.1128/AAC.02782-14.
- Cubillos-Ruiz A, Alcantar MA, Donghia NM, Cárdenas P, Avila-Pacheco J, Collins JJ. An engineered live biotherapeutic for the prevention of antibiotic-induced dysbiosis. Nat Biomed Eng. 2022;6(7):910–921. doi:10.1038/s41551-022-00871-9.
- Mensink MA, Frijlink HW, van der Voort Maarschalk K, Hinrichs WLJ. Inulin, a flexible oligosaccharide I: review of its physicochemical characteristics. Carbohydr Polym. 2015;130:405–419. doi:10.1016/j.carbpol.2015.05.026.
- Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio. 2019;10(1):e02566–18. doi:10.1128/mBio.02566-18.
- Chijiiwa R, Hosokawa M, Kogawa M, Nishikawa Y, Ide K, Sakanashi C, Takahashi K, Takeyama H. Single-cell genomics of uncultured bacteria reveals dietary fiber responders in the mouse gut microbiota. Microbiome. 2020;8(1). doi:10.1186/s40168-019-0779-2.
- Le Bastard Q, Chapelet G, Javaudin F, Lepelletier D, Batard E, Montassier E. The effects of inulin on gut microbial composition: a systematic review of evidence from human studies. Eur J Clin Microbiol Infect Dis. 2020;39(3):403–413. doi:10.1007/s10096-019-03721-w.
- Gregoire M, Berteau F, Bellouard R, Lebastard Q, Aubert P, Gonzales J, Javaudin F, Bessard A, Bemer P, Batard É. et al. A murine model to study the gut bacteria parameters during complex antibiotics like cefotaxime and ceftriaxone treatment. Comput Struct Biotechnol J. 2021;19:1423–1430. doi:10.1016/j.csbj.2021.02.019.
- Zhu L, Qin S, Zhai S, Gao Y, Li L. Inulin with different degrees of polymerization modulates composition of intestinal microbiota in mice. FEMS Microbiol Lett. 2017;364(10). doi:10.1093/femsle/fnx075.
- Rodriguez J, Hiel S, Neyrinck AM, Le Roy T, Pötgens SA, Leyrolle Q, Pachikian BD, Gianfrancesco MA, Cani PD, Paquot N. et al. Discovery of the gut microbial signature driving the efficacy of prebiotic intervention in obese patients. Gut. 2020;69(11):1975–1987. doi:10.1136/gutjnl-2019-319726.
- Caballero S, Kim S, Carter RA, Leiner IM, Sušac B, Miller L, Kim GJ, Ling L, Pamer EG. Cooperating commensals restore colonization resistance to vancomycin-resistant Enterococcus faecium. Cell Host Microbe. 2017;21(5):592–602.e4. doi:10.1016/j.chom.2017.04.002.
- Juhász J, Ligeti B, Gajdács M, Makra N, Ostorházi E, Farkas FB, Stercz B, Tóth Á, Domokos J, Pongor S. et al. Colonization Dynamics of Multidrug-Resistant Klebsiella pneumoniae Are Dictated by Microbiota-Cluster Group Behavior over Individual Antibiotic Susceptibility: A Metataxonomic Analysis. Antibiotics (Basel). 2021;10(3):268. doi:10.3390/antibiotics10030268.
- Wang L, Wang Z, Lan Y, Tuo Y, Ma S, Liu X. Inulin attenuates blood–brain barrier permeability and alleviates behavioral disorders by modulating the TLR4/MyD88/NF-κB pathway in mice with chronic stress. J Agric Food Chem. 2023;71(36):13325–13337. doi:10.1021/acs.jafc.3c03568.
- Hoving LR, Katiraei S, Pronk A, Heijink M, Vonk KKD, Amghar-el Bouazzaoui F, Vermeulen R, Drinkwaard L, Giera M, van Harmelen V. et al. The prebiotic inulin modulates gut microbiota but does not ameliorate atherosclerosis in hypercholesterolemic APOE*3-Leiden.CETP mice. Sci Rep. 2018;8(1):16515. doi:10.1038/s41598-018-34970-y.
- Le Guern R, Grandjean T, Stabler S, Bauduin M, Gosset P, Kipnis É, Dessein R. Gut colonisation with multidrug-resistant Klebsiella pneumoniae worsens Pseudomonas aeruginosa lung infection. Nat Commun. 2023;14(1):78. doi:10.1038/s41467-022-35767-4.
- Stiefel U, Rao A, Pultz MJ, Jump RLP, Aron DC, Donskey CJ. Suppression of gastric acid production by proton pump inhibitor treatment facilitates colonization of the large intestine by vancomycin-resistant Enterococcus spp. And Klebsiella pneumoniae in clindamycin-treated mice. Antimicrob Agents Chemother. 2006;50(11):3905–3907. doi:10.1128/AAC.00522-06.
- Ishnaiwer M, Bezabih Y, Javaudin F, Sassi M, Bemer P, Batard E, Dion M. In vitro and in vivo activity of new strains of Bacillus subtilis against ESBL-producing Escherichia coli: an experimental study. J Appl Microbiol. 2022;132(3):2270–2279. doi:10.1111/jam.15329.
- Danylec N, Stoll DA, Göbl A, Huch M, Rasko D. Draft genome sequences of 13 isolates of adlercreutzia equolifaciens, Eggerthella lenta, and gordonibacter urolithinfaciens, isolated from human fecal samples in Karlsruhe, Germany. Microbiol Resour Announc. 2020;9(8):e00017–20. doi:10.1128/MRA.00017-20.
- Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi:10.1093/bioinformatics/bts565.
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 2012;13(1):134. doi:10.1186/1471-2105-13-134.
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Lu J, Breitwieser FP, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci. 2017;3:e104. doi:10.7717/peerj-cs.104.
- Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):257. doi:10.1186/s13059-019-1891-0.
- Beresford-Jones BS, Forster SC, Stares MD, Notley G, Viciani E, Browne HP, Boehmler DJ, Soderholm AT, Kumar N, Vervier K. et al. The mouse gastrointestinal bacteria catalogue enables translation between the mouse and human gut microbiotas via functional mapping. Cell Host Microbe. 2022;30(1):124–138.e8. doi:10.1016/j.chom.2021.12.003.
- Yin X, Jiang X-T, Chai B, Li L, Yang Y, Cole JR, Tiedje JM, Zhang T. Args-OAP v2.0 with an expanded SARG database and hidden Markov models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics. 2018;34(13):2263–2270. doi:10.1093/bioinformatics/bty053.
- Beghini F, McIver LJ, Blanco-Míguez A, Dubois L, Asnicar F, Maharjan S, Mailyan A, Manghi P, Scholz M, Thomas AM. et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife. 2021;10:e65088. doi:10.7554/eLife.65088.
- Caspi R, Billington R, Keseler IM, Kothari A, Krummenacker M, Midford PE, Ong WK, Paley S, Subhraveti P, Karp PD. et al. The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res. 2020;48(D1):D445–D453. doi:10.1093/nar/gkz862.
- Drula E, Garron M-L, Dogan S, Lombard V, Henrissat B, Terrapon N. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 2022;50(D1):D571–D577. doi:10.1093/nar/gkab1045.
- Moaligou C, Dion M, Ishnaiwer M, Dailly É, Batard É, Javaudin F. Pantoprazole promotes sustained intestinal carriage of multidrug-resistant Escherichia coli in amoxicillin-treated mice. J Appl Microbiol lxad. 2023;223(10). doi:10.1093/jambio/lxad223.
- Ni W, Cai X, Liang B, Cai Y, Cui J, Wang R. Effect of proton pump inhibitors on in vitro activity of tigecycline against several common clinical pathogens. PLOS ONE. 2014;9(1):e86715. doi:10.1371/journal.pone.0086715.
- van den Bunt G, van Pelt W, Hidalgo L, Scharringa J, de Greeff SC, Schürch AC, Mughini-Gras L, Bonten MJM, Fluit AC. Prevalence, risk factors and genetic characterisation of extended-spectrum beta-lactamase and carbapenemase-producing enterobacteriaceae (ESBL-E and CPE): a community-based cross-sectional study, the Netherlands, 2014 to 2016. Euro Surveill. 2019;24(41):1800594. doi:10.2807/1560-7917.ES.2019.24.41.1800594.
- Willems RPJ, van Dijk K, Ket JCF, Vandenbroucke-Grauls CMJE. Evaluation of the association between gastric acid suppression and risk of intestinal colonization with multidrug-resistant microorganisms: a systematic review and meta-analysis. JAMA Internal Medicine. 2020;180(4):561–571. doi:10.1001/jamainternmed.2020.0009.
- Raffelsberger N, Buczek DJ, Svendsen K, Småbrekke L, Pöntinen AK, Löhr IH, Andreassen LLE, Simonsen GS, Sundsfjord A, Gravningen K. et al. Community carriage of ESBL-producing Escherichia coli and Klebsiella pneumoniae: a cross-sectional study of risk factors and comparative genomics of carriage and clinical isolates. mSphere. 2023;8(4):e0002523. doi:10.1128/msphere.00025-23.
- Le Bastard Q, Berthelot L, Soulillou J-P, Montassier E. Impact of non-antibiotic drugs on the human intestinal microbiome. Expert Rev Mol Diagn. 2021;21(9):911–924. doi:10.1080/14737159.2021.1952075.
- O’Donoghue C, Solomon K, Fenelon L, Fitzpatrick F, Kyne L. Effect of proton pump inhibitors and antibiotics on the gut microbiome of hospitalised older persons. J Infect. 2016;72(4):498–500. doi:10.1016/j.jinf.2016.01.015.
- Liu W, Li X, Zhao Z, Pi X, Meng Y, Fei D, Liu D, Wang X. Effect of chitooligosaccharides on human gut microbiota and antiglycation. Carbohyd Polym. 2020;242:116413. doi:10.1016/j.carbpol.2020.116413.
- Chung YW, Gwak H-J, Moon S, Rho M, Ryu J-H, Dong Q. Functional dynamics of bacterial species in the mouse gut microbiome revealed by metagenomic and metatranscriptomic analyses. PloS One. 2020;15(1):e0227886. doi:10.1371/journal.pone.0227886.
- Smith BJ, Miller RA, Schmidt TM, Suen G. Muribaculaceae genomes assembled from metagenomes suggest genetic drivers of differential response to acarbose treatment in mice. mSphere. 2021;6(6):e0085121. doi:10.1128/msphere.00851-21.
- Vacca M, Celano G, Calabrese FM, Portincasa P, Gobbetti M, De Angelis M. The controversial role of human gut lachnospiraceae. Microorganisms. 2020;8(4):E573. doi:10.3390/microorganisms8040573.
- Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A. The genus alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol. 2020;11:906. doi:10.3389/fimmu.2020.00906.
- Ormerod KL, Wood DLA, Lachner N, Gellatly SL, Daly JN, Parsons JD, Dal’molin CGO, Palfreyman RW, Nielsen LK, Cooper MA. et al. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome. 2016;4(1). doi:10.1186/s40168-016-0181-2.
- Wu G, Zhang C, Wang J, Zhang F, Wang R, Shen J, Wang L, Pang X, Zhang X, Zhao L. et al. Diminution of the gut resistome after a gut microbiota-targeted dietary intervention in obese children. Sci Rep. 2016;6(1):24030. doi:10.1038/srep24030.
- Montassier E, Valdés-Mas R, Batard E, Zmora N, Dori-Bachash M, Suez J, Elinav E. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat Microbiol. 2021;6(8):1043–1054. doi:10.1038/s41564-021-00920-0.
- Mayo B, Vázquez L, Flórez AB. Equol: a bacterial metabolite from the Daidzein Isoflavone and its presumed beneficial health effects. Nutrients. 2019;11(9):E2231. doi:10.3390/nu11092231.
- Yu Z-T, Yao W, Zhu W-Y. Isolation and identification of equol-producing bacterial strains from cultures of pig faeces. FEMS Microbiol Lett. 2008;282(1):73–80. doi:10.1111/j.1574-6968.2008.01108.x.
- Liang W, Zhao L, Zhang J, Fang X, Zhong Q, Liao Z, Wang J, Guo Y, Liang H, Wang L. et al. Colonization potential to reconstitute a microbe community in pseudo germ-free mice after fecal microbe transplant from equol Producer. Front Microbiol. 2020;11:1221. doi:10.3389/fmicb.2020.01221.

