Abstract
Geographic isolation and colonization events have been considered complimentary historical processes in shaping the contemporary structure of regional biotas and taxonomic diversity. Accordingly, insight regarding vicariance and geodispersal might be gained by the analysis of phylogenetic pattern of related but geographically isolated taxa. Here, we examined endangered Rhodeus pseudosericeus in the Korean Peninsula based on mitochondrial cytochrome b sequences to provide information regarding the evolutionary origin of Amur-European bitterlings, Rhodeus sericeus and Rhodeus amarus. Our data and the contemporary distribution of R. pseudosericeus indicate that founding individuals have colonized the western coast of the Korean Peninsula via the paleo-Huang He River and since been subdivided into populations isolated by drainage formation. Our phylogenetic analyses indicated that it was likely R. pseudosericeus had a sister relationship to the R. sericeus–amarus complex, indicating that the R. sericeus–amarus clade originated from the dispersal of either R. pseudosericeus or its ancestral lineage. The geodispersal scenario based on our phylogenetic analyses supports the previous hypothesis that the Amur River likely created confluences with some tributaries of the paleo-Huang He River. Overall, the present study offers new insight into the taxonomic entity of R. pseudosericeus and a more comprehensive understanding of the phylogeographic history of R. sericeus and European bitterling lineages.
Introduction
Acheilognathinae (bitterlings) is a taxonomic subfamily of approximately 60 small freshwater fish species that are divided into three genera: Acheilognathus, Tanakia, and Rhodeus (Arai & Akai Citation1988; Nelson Citation2006; Kitamura et al. Citation2012). Over the last several decades, this subfamily has attracted scientific interest among evolutionary biologists due to the unique breeding behavior of its members, which includes obligate spawning in the gill cavities of living freshwater unionid and margaritiferid mussels (Smith et al. Citation2004; Spence & Smith Citation2013). The majority of bitterling species are found in lotic and lentic freshwater systems throughout East Asia (Nelson Citation2006), though some are currently endangered (Ohta et al. Citation2001; Kubota & Watanabe Citation2003; Kitamura et al. Citation2009).
A single valid species, Rhodeus amarus, is present throughout Europe (Reichard et al. Citation2007; Zaki et al. Citation2008; Bryja et al. Citation2010). A phylogeographical study based on a cytochrome b (cyt b) gene indicated that there are four lineages of R. amarus (; Bohlen et al. Citation2006; Bryja et al. Citation2010), among which two were recently distinguished from R. amarus as separate species, Rhodeus meridionalis (from the Vardar River in Greece; Bohlen et al. Citation2006) and Rhodeus colchicus (from an isolated region in central Caucasus; Bohlen et al. Citation2006). All four lineages have traditionally been identified as R. sericeus, which is native to northeast Asia (Holcík & Jedlicka Citation1994), or as a subspecies, R. s. amarus, until they were validated as discrete species by taxonomic reevaluation (Bohlen et al. Citation2006). The cyt b gene has also demonstrated that the common ancestor for the four European lineages originated from the dispersal of R. sericeus (; Bohlen et al. Citation2006).
Rhodeus pseudosericeus, an endemic species in the Korean Peninsula (), was also identified as R. sericeus when it was first recovered (Chae & Yang Citation1993). Despite their morphological similarities, R. pseudosericeus is clearly distinct from R. sericeus as well as from the European R. amarus lineages based on several characteristics, justifying its status as a separate species (Arai et al. Citation2001). This species is currently classified as endangered under the Protection of Wild Fauna and Flora Act of the Korean Ministry of Environment. Our study was designed to provide novel data for the relationship between the R. pseudosericeus and R. sericeus–amarus complex and their phylogenetic placement in the subfamily. Given that they are recovered as a monophyletic clade, R. pseudosericeus may provide a critical model system for the investigation of the origin and historical dispersal of R. sericeus and European bitterling lineages. Two exclusively alternative dispersal scenarios could be envisaged (); namely, either R. pseudosericeus was colonized by R. sericeus or its ancestral lineage or R. sericeus originated from the dispersal of R. pseudosericeus or its ancestral lineage.
Materials and methods
Sampling and sequencing
R. pseudosericeus is only found in three western-flowing drainages in the Korean Peninsula (). For this study, individuals were collected during March–September of 2011 from six localities (; ). Collection and preservation were conducted under the permission of the local administrative institutions of the Ministry of Environment in South Korea. Total genomic DNA was extracted from caudal peduncle tissue using the Wizard Genomic DNA purification kit (PROMEGA, Fitchburg, WI, USA) in accordance with the manufacturer's protocol. The mitochondrial cyt b gene was used for the analysis of genetic variation within and among populations. Each amplification was conducted in a 50-µL reaction volume composed of ∼75 ng genomic DNA extract, 0.25 mM of each deoxynucleotide, 0.4 µM of each primer (forward: Brito et al. Citation1997; backward: Perdices et al. Citation2002), 3 mM MgCl2, 1× PCR buffer, and 0.25 units of Taq DNA polymerase (SOLGENT, Daejeon, South Korea). GenePro (BIOER, Hangzhou, P.R. China) was used to amplify the cyt b gene at an annealing temperature of 54°C. Amplified products were directly sequenced in both directions using the PCR primers with an ABI PRISM BigDye terminator system in an ABI3700 automatic sequencer (GENOTECH, Daejeon, South Korea).
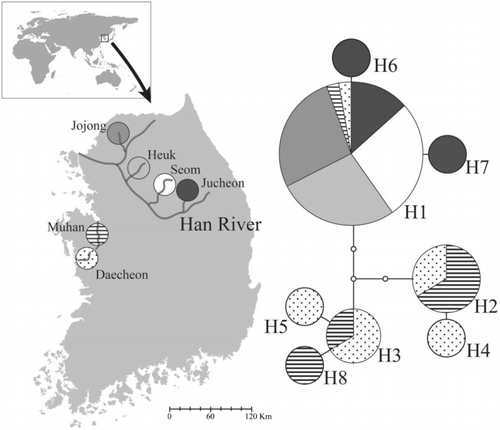
Table 1. List of the six R. pseudosericeus populations used in the present study.
Data analysis
The nucleotide sequences of the cyt b gene were confirmed through Basic Local Alignment Search Tool (BLAST) searches, edited with GENEIOUS, ver. 5.4, and aligned with the nucleotide sequences from other bitterling species and out-group taxa () retrieved from GenBank in the program MEGA, ver. 5.1 (Tamura et al. Citation2011). Genetic diversity at the intrapopulation level was quantified by estimating the number of haplotypes, haplotype diversity (H d), and nucleotide diversity (π) using DnaSP, ver. 5.10 (Librado & Rozas Citation2009). An un-rooted network was constructed to depict the relationships among haplotypes based on the 95% statistical parsimony method using TCS, ver. 1.21 (Clement et al. Citation2000). Genetic structure was estimated with a hierarchical analysis of molecular variance (AMOVA) implemented in ARLEQUIN, ver. 3.1 (Excoffier et al. Citation2005) to examine whether all populations were regarded as a single group ().
Table 2. GenBank accession numbers and the references of the haplotypes used in the BEAST species-tree reconstruction based on fossil calibration at the stem of Acheilognathinae lineage and a substitution rate for the cyt b gene ().
Table 3. Summary of the AMOVA results that partition genetic variation among six R. pseudosericeus populations from the Korean Peninsula to two different geographic levels (among groups: the Han River vs. Muhan-Daechon).
For the phylogenetic analysis, out-group taxa included Tanichthys albonubes from other subfamily within Cyprinidae and Kichulchoia brevifasciata from Cobitidae, Cypriniformes (). The GTR+I+G model was selected as the most appropriate nucleotide substitution model with MrModeltest, ver. 2.3 (Nylander Citation2004). Phylogenetic relationship and divergent time estimation were inferred in BEAST, ver. 1.7.4 (Drummond et al. Citation2012), enforcing a relaxed molecular clock under models of uncorrelated lognormal distribution rate and a Yule speciation. Two independent runs of 1 × 108 generations were conducted, sampling parameters every 1000 generations and utilizing the fossil calibration point of 23 Mya (early Miocene) at the stem of Acheilognathinae lineage (Yang et al. Citation2011) and a cyt b substitution rate of 0.76% (Zardoya & Doadrio Citation1999). Lognormal distributions were applied for the fossil calibration point with a mean of 23.0, SD of 0.2, and an offset of 0.0, and for the rate prior for cyt b with a mean of 0.76, SD of 0.49, and an offset of 0.0. LOGCOMBINER, ver. 1.7.4 (Drummond et al. Citation2012), was used to combine two independent BEAST analyses, which converged on the same parameter space. The tree was annotated with TREEANNOTATOR, ver. 1.7.4 (Drummond et al. Citation2012) and was visualized in FIGTREE, ver. 1.4.0.
Results
In the present study, cyt b gene sequences of 860 nucleotides from 60 individuals yielded eight distinct haplotypes (deposited in GenBank under accession nos. KJ028096–028103; ), with one of them (H1) occupying a wide geographic distribution (). The remaining haplotypes were of low frequency, with each one represented by just one or two populations (). Three populations (Heuk, Jojong, and Seom) from the Han drainage harbored only a single haplotype, H1 (). Only seven variable nucleotide sites were found among 860 bp, as were the corresponding haplotype diversity (Hd = 0.512) and nucleotide diversity (π = 0.0022). Two distinct genetic clusters, the Han River populations and Muhan-Daechon, were identified from the minimum parsimony network (), and the results of AMOVA indicated that more genetic variance occurred among clusters than among populations or individuals within populations ().
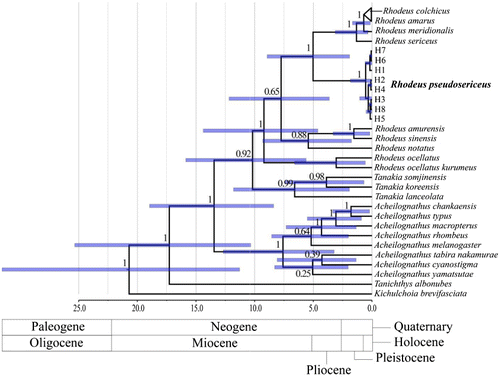
In our phylogenetic analysis, R. sericeus and R. amarus lineages formed a monophyletic group. Eight haplotypes of R. pseudosericeus were recovered as the most likely sister group of R. sericeus–amarus clade by a strong statistical value (), indicating that the R. sericeus and European bitterlings originated from the dispersal of either R. pseudosericeus or its ancestral lineage or vice versa. The three genera of Acheilognathinae formed complete and independent monophyletic clades, with Acheilognathus being placed at the ancestral position (). Tanakia was recovered as a sister to the monophyly of Rhodeus species (). Within Rhodeus, the remaining species were basal and sister to R. pseudosericeus plus R. sericeus–amarus complex with high statistical supports (). The root nodes for Acheilognathinae and Rhodeus were estimated to be about 17.27 Mya (CI: 10.32–25.33) and 9.20 Mya (4.60–14.37), respectively. An age estimate of 5.02 Mya (1.90–8.94) was assigned to the node leading to the separation between the R. pseudosericeus and R. sericeus–amarus complex ().
Discussion
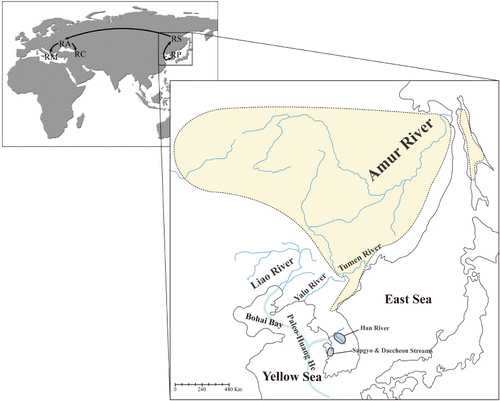
Low to moderate levels of intrapopulation genetic diversity were shown across the six populations investigated. Surprisingly, more diverse haplotypes were found from the much smaller Daecheon and Sapgyo streams than those observed from the Han River, one of the major freshwater systems in the Korean Peninsula. The current distribution of R. pseudosericeus and the population genetic structure may bear the mark of drainage formation and colonization dating back to when sea levels were much lower than at present. It is well known that the Huang He River, which is the major drainage system running through China and discharging eastward into the Yellow Sea, likely created confluences with the present-day western-flowing drainages along the western coast of the Korean Peninsula during the Quaternary glacial periods (; Nishimura Citation1974). Thus, it is conceivable that a refugial unit of R. pseudosericeus (or its lineage) has colonized along the current western coastal areas of the Korean Peninsula via the paleo-Huang He River and has been subdivided into populations that were isolated by the disappearance of paleo-river systems and drainage formation. Following colonization events, many populations of R. pseudosericeus might have been depleted, which could be attributed to the contemporary scattered and limited distribution. Alternatively, the Han drainage and Daecheon and Sapgyo streams may have been independently colonized by different founding populations, which might have led to the genetic subdivision between the Han River and Daecheon-Muhan group. In addition to geographic isolation and colonization patterns, the genetic structuring might be at least partially due to genetic drift arising from repeated population bottlenecks.
Insight regarding geodispersal processes of R. sericeus and R. amarus lineages could be obtained by our analyses of the relationship with R. pseudosericeus and the phylogenetic position within Acheilognathinae. While R. sericeus and R. amarus lineages formed a monophyletic group, R. pseudosericeus could be treated as a sister of the R. sericeus–amarus complex by strong node-supporting values. Given that most of Rhodeus bitterlings are currently distributed in the paleo-Huang He River with being placed at the basal position in the phylogenetic tree, our results indicated that the R. sericeus–amarus complex was derived from the dispersal of either R. pseudosericeus or its ancestral lineage. Solely based on this scenario, it is conceivable that the founding individuals of R. sericeus originated from the paleo-Huang He River colonized the Amur River. However, the paleo-Huang He River might not appear in the present Yellow Sea area at the estimated time of divergence between R. pseudosericeus and R. sericeus–amarus complex, since no glacial periods have been required around that time. Those two phylogenetic groups might have been differentiated from each other before their dispersal into the Amur region.
It is unclear how this bitterling dispersed from the paleo-Huang He River to the geographically isolated Amur River. The Amur River likely created confluences with the Liao River, a tributary of the paleo-Huang He River (), as those two rivers contain freshwater fauna with a high degree of similarity (Sakai et al. Citation2006). More detailed inferences regarding their processes of speciation and colonization require additional extensive sampling of R. sericeus and the related species throughout their distribution range. In addition, the more complete resolution of the colonization history suggested in the present study could be obtained by the investigation of other bitterling species. For example, Rhodeus amurensis, native to the Amur River, is closely related to Rhodeus sinensis occurring in the rivers originated from the paleo-Huang He River (see also ).
Gil et al. (Citation2007) reported that R. sericeus individuals occurred in eastern-flowing freshwater systems – probably originated from the paleo-Amur River – from North Korea. In addition, Arai et al. (Citation2001) investigated some North Korean R. sericeus samples based on morphological characteristics and found the overall similarity to R. pseudosericeus. Considering that R. pseudosericeus populations exist along the eastern coast in North Korea, it may also be possible to infer that the divergence between R. pseudosericeus and R. sericeus–amarus complex had happened in the Amur region and the R. pseudosericeus populations examined in the present study had been colonized by North Korean populations. From this idea, however, it should be proved how these two geographically separated rivers could have created confluences via the Korean Peninsula. Direct comparison with North Korean samples may provide more concrete solution for the origin and the historical dispersal pattern of R. pseudosericeus.
Taken together, the present study provides new insight into the taxonomic entity of R. pseudosericeus and more comprehensive understanding of the phylogeographic history of R. sericeus and European bitterlings. In summary, our results indicated that (1) founding individuals of R. pseudosericeus have colonized the western coast of the Korean Peninsula via the paleo-Huang He River, (2) R. sericeus and R. amarus shared the common ancestor with R. pseudosericeus or might originate from the dispersal of its ancestral lineage, and (3) the Amur River likely created confluences with the paleo-Huang He River.
Acknowledgment
We are grateful to Seong Jang Jo, Akimitsu Hanado, and Jin Young Chu for their helpful support in the field, and Daemin Kim for his assistance with phylogenetic analyses. We also thank two anonymous reviewers for helpful comments on the manuscript.
Funding
This project was supported by the grants from the Korean National Institute of Biological Resources [grant number 074-1800-1844-304] and the Korean National Research Foundation [grant number NRF-2011-0014704).
Additional information
Funding
References
- Arai R, Akai Y. 1988. Acheilognathus melanogaster, a senior synonym of A. moriokae, with a revision of the genera of the subfamily Acheilognathinae (Cypriniformes, Cyprinidae). Bull Nat Sci Mus Tokyo Ser A. 14:199–213.
- Arai R, Jeon S-R, Ueda T. 2001. Rhodeus pseudosericeus sp. nov., a new bitterling from South Korea (Cyprinidae, Acheilognathinae). Ichthyol Res. 48:275–282.10.1007/s10228-001-8146-1
- Bohlen J, Šlechtová V, Bogutskaya N, Freyhof J. 2006. Across Siberia and over Europe: phylogenetic relationships of the freshwater fish genus Rhodeus in Europe and the phylogenetic position of R. sericeus from the River Amur. Mol Phylogenet Evol. 40:856–865.10.1016/j.ympev.2006.04.020
- Brito RM, Briolay J, Galtier N, Bouvet Y, Coelho MM. 1997. Phylogenetic relationships within genus Leuciscus (Pisces, Cyprinidae) in Portuguese fresh waters, based on mitochondrial DNA cytochrome b sequences. Mol Phylogenet Evol. 8:435–442.10.1006/mpev.1997.0429
- Bryja J, Smith C, Konecný A, Reichard M. 2010. Range-wide population genetic structure of the European bitterling (Rhodeus amarus) based on microsatellite and mitochondrial DNA analysis. Mol Ecol. 19:4708–4722.10.1111/j.1365-294X.2010.04844.x
- Chae BS, Yang HJ. 1993. A new distributional record of an Acheilognathine fish, Rhodeus sericeus (Pallas) (Cyprinidae, Pisces) in South Korea. Korean J Zool. 36:175–180. Korean, English abstract.
- Clement M, Posada D, Crandall KA. 2000. TCS: a computer program to estimate gene genealogies. Mol Ecol. 9:1657–1659.10.1046/j.1365-294x.2000.01020.x
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the beast 1.7. Mol Biol Evol. 29:1969–1973.10.1093/molbev/mss075
- Excoffier L, Laval G, Schneider S. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinf Online. 1:47–50.
- Fang F, Noren M, Liao TY, Kallersjo M, Kullander SO. 2009. Molecular phylogenetic interrelationships of the south Asian cyprinid genera Danio, Devario and Microrasbora (Teleostei, Cyprinidae, Danioninae). Zool Scr. 38:237–256.10.1111/j.1463-6409.2008.00373.x
- Gil JW, Hong YP, Kim S. 2007. Fish fauna in southern river of Bukcheong and Brackish lakes, the Shinpo District, North Korea. Korean J Environ Biol. 25:279–287. Korean, English abstract.
- Holcík J, Jedlicka L. 1994. Geographical variation of some taxonomically important characters in fishes: the case of the bitterling Rhodeus sericeus. Environ Biol Fish. 41:147–170.10.1007/BF02197842
- Hwang D-S, Byeon HK, Lee J-S. 2014. Complete mitochondrial genome of the freshwater fish, Acheilognathus somjinensis (Cypriniformes, Cyprinidae). Mitochondrial DNA. 25:13–14.10.3109/19401736.2013.775266
- Hwang D-S, Lee W-O, Lee J-S. 2013. Complete mitochondrial genome of the Korean bitterling Acheilognathus koreensis (Cypriniformes; Cyprinidae). Mitochondrial DNA. 24:414–415.10.3109/19401736.2013.766178
- Hwang D-S, Lee W-O, Lee J-S. 2014. Complete mitochondrial genome of the freshwater fish, Rhodeus suigensis (Cypriniformes, Cyprinidae). Mitochondrial DNA. 25:5–6.10.3109/19401736.2013.775262
- Kim D, Conway KW, Jeon H-B, Kwon Y-S, Won Y-J. 2013. High genetic diversity within the morphologically conservative Dwarf loach, Kichulchoia brevifasciata (Teleostei: Cobitidae), an endangered freshwater fish from South Korea. Conserv Genet. 14:757–769.10.1007/s10592-013-0462-2
- Kitamura J, Nagata N, Nakajima J, Sota T. 2012. Divergence of ovipositor length and egg shape in a brood parasitic bitterling fish through the use of different mussel hosts. J Evol Biol. 25:566–573.10.1111/j.1420-9101.2011.02453.x
- Kitamura J, Negishi JN, Nishio M, Sagawa S, Akino J, Aoki S. 2009. Host mussel utilization of the Itasenpara bitterling (Acheilognathus longipinnis) in the Moo River in Himi, Japan. Ichthyol Res. 56:296–300.10.1007/s10228-008-0082-x
- Kubota H, Watanabe K. 2003. Genetic diversity in wild and reared populations of the Japanese bitterling Tanakia tanago (Cyprinidae). Ichthyol Res. 50:123–128.10.1007/s10228-002-0146-2
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452.10.1093/bioinformatics/btp187
- Nelson JS. 2006. Fishes of the world. New York: Wiley and Sons.
- Nishimura S. 1974. Origin of the sea of Japan–approach from biogeography. Tokyo: Tsukiji Shokan. Japanese.
- Nylander JAA. 2004. MrModeltest v2 [software]. Uppsala: Evolutionary Biology Centre, Uppsala University. Distributed by the author.
- Ohta H, Kawamura K, Unuma T, Takegoshi Y. 2001. Cryopreservation of the sperm of the Japanese bitterling. J Fish Biol. 58:670–681.10.1111/j.1095-8649.2001.tb00521.x
- Perdices A, Bermingham E, Montilla A, Doadrio I. 2002. Evolutionary history of the genus Rhamdia (Teleostei: Pimelodidae) in Central America. Mol Phylogenet Evol. 25:172–189.10.1016/S1055-7903(02)00224-5
- Reichard M, Przybylski M, Kaniewska P, Liu H, Smith C. 2007. A possible evolutionary lag in the relationship between freshwater mussels and European bitterling. J Fish Biol. 70:709–725.10.1111/j.1095-8649.2007.01333.x
- Saitoh K, Sado T, Mayden RL, Hanzawa N, Nakamura K, Nishida M, Miya M. 2006. Mitogenomic evolution and interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): the first evidence toward resolution of higher-level relationships of the world's largest freshwater fish clade based on 59 whole mitogenome sequences. J Mol Evol. 63:826–841.10.1007/s00239-005-0293-y
- Sakai H, Ito Y, Shedko SV, Safronov SN, Frolov SV, Chereshnev IA, Jeon SR, Goto A. 2006. Phylogenetic and taxonomic relationships of northern Far Eastern phoxinin minnows, Phoxinus and Rhynchocypris (Pisces, Cyprinidae), as inferred from allozyme and mitochondrial 16S rRNA sequence analyses. Zool Sci. 23:323–331.10.2108/zsj.23.323
- Smith C, Reichard M, Jurajda P, Przybylski M. 2004. The reproductive ecology of the European bitterling (Rhodeus sericeus). J Zool. 262:107–124.10.1017/S0952836903004497
- Spence R, Smith C. 2013. Rose bitterling (Rhodeus ocellatus) embryos parasitize freshwater mussels by competing for nutrients and oxygen. Acta Zool. 94:113–118.10.1111/j.1463-6395.2011.00532.x
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.10.1093/molbev/msr121
- Yang Q, Zhu Y, Xiong B, Liu H. 2011. Acheilognathus changtingensis sp. nov., a new species of the cyprinid genus Acheilognathus (Teleostei: Cyprinidae) from Southeastern China based on morphological and molecular evidence. Zool Sci. 28:158–167.10.2108/zsj.28.158
- Zaki SAH, Jordan WC, Reichard M, Przybylski M, Smith C. 2008. A morphological and genetic analysis of the European bitterling species complex. Biol J Linn Soc. 95:337–347.10.1111/j.1095-8312.2008.01050.x
- Zardoya R, Doadrio I. 1999. Molecular evidence on the evolutionary and biogeographical patterns of European cyprinids. J Mol Evol. 49:227–237.10.1007/PL00006545

