?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Background: TIMP3 is a multifunctional proteolytic enzyme belonging to TIMPs family and acts as a potent inhibitor of matrix metalloproteinases (MMPs). TIMP3 possesses a tumor suppresive function by directly promoting tumor cell apoptosis, preventing angiogenesis and extracellular matrix remodelling. The lower expression of TIMP3 was associated with poor prognosis and overall survival in various cancer types. The aim of this study was to evaluate the association of TIMP3 protein expression with ovarian cancer (OC) clinicopathological features and survival outcomes.Patients and Methods: One hundred forty four of OC FFPE samples were collected from King Abdulaziz University Hospital, Saudi Arabia and constructed in tissue microarray (TMA) slides. Automated Ventana immunostainer platform was used to evaluate TIMP3 protein expression patterns.Results: The study showed that TIMP3 exhibits cytoplasmic localisation. This TIMP3 protein expression was not associated with age, tumor size and the involvement of lymph nodes (p > 0.05). However, it was significantly correlated with tumor stage (p < 0.05) and borderline significant with endpoint status (p = 0.07). Interestingly, the Kaplan-Meier analysis of disease specific survival (DSS) outcomes showed a significant association (p = 0.02, log rank) between OC patients with higher TIMP3 expression compared to those with lower expression. In fact, OC patients with high TIMP3 expression had longer survivals. Multivariate Cox’s regression analysis suggests that low TIMP3 protein expression pattern is an independent poor survival marker (p = 0.025).Conclusion: Cytoplasmic TIMP3 protein expression could be used as a good prognosticator to stratify poorly prognostic OC patients in order to personlaize their disease management.
1. Introduction
Ovarian cancer (OC) has a high mortality rate due to late presentation to the clinic and more resistance to conventional chemotherapy. This is likely to be attributed to its unique pathophysiological features and molecular heterogeneity [Citation1]. The worldwide incidence rate of OC varies significantly depending upon multiple risk factors such as early-age menarche, delayed menopause, elderly age, higher number of ovulatory cycles, nulliparity, and use of fertility drugs [Citation2–5]. The epithelial ovarian cancer (EOC) is the most frequent subtype accounting for approximately 90% of OC cases [Citation6,Citation7]. In Saudi Arabia, OC accounted for 3.3% of all cases of newly diagnosis female patients with an average of Age-Standardized Incidence Rate (ASR) of 3 per 100,000 [Citation8]. The 5-year survival rate among Saudi patients with advanced stages (III or IV) was very low (15–29%), but it is far better (88%) for women diagnosed at early stage (I). The majority of OC patients are diagnosed at advanced stages with metastatic dissemination to peritoneal cavity and distant organs where the therapeutic options are unfortunately very limited and requires stringent multimodal therapy modules [Citation9]. Usually, patients respond positively to the treatment, but the majority of the patients develop resistance to chemotherapy leading to the relapsed fatal disease.
Besides the aberrant genomic and epigenetic changes in ovarian epithelial cells [Citation10,Citation11], mutations in genes responsible for maintaining normal ovarian epithelial tissue architecture can also promote EOC progression [Citation12,Citation13]. Before metastasis, epithelial cells acquired mesenchymal characteristics through a process called epithelial-to-mesenchymal transition (EMT) followed by the dissolution of extracellular matrix (ECM) to create suitable environment for cell detachment, cell invasion and migration [Citation14,Citation15]. The physiological degradation of ECM is mainly mediated by specific metal ion containing proteolytic enzymes called the matrix metalloproteinases (MMPs) [Citation16–18]. These MMPs cleave the ECM to accelerate tissue remodeling that promotes metastasis-related processes including cells signaling and communication [Citation19], cell growth, invasion and migration. The function of the MMPs is regulated and controlled by their natural inhibitors called tissue inhibitors of metallopeptidase (TIMPs). Published data revealed that the imbalance between MMPs and TIPMs has been involved in the development of several malignancies [Citation3,Citation9,Citation13,Citation14]. In particular, TIPM3 is a secreted glycoprotein of the TIMP family. But unlike other TIMPs, TIMP3 is the only MMPs inhibitor know so far to be incorporated into the ECM. Impaired TIMP3 level has been reported in many cancers [Citation20]. However, TIMP3 increased expression was associated with good prognosis indicating a potential role as a tumor suppressor and/or biomarker [Citation21,Citation22]. However, little is known about the expression of TIMP3 in OC. The current study was undertaken to evaluate the correlation between TIMP3 expression and the clinico-pathological features of Saudi OC patients and to assess its prognostic value in order to offer more effective OC management biomarkers.
2. Patients and methods
2.1. Patients
Tissue microarrays (TMAs) were constructed from formalin-fixed paraffin-embedded (FFPE) tissue of OC patients who underwent surgery at the Departments of Pathology & Gynecology, King Abdulaziz University Hospital (KAUH) in the period between 1995 and 2014 after obtaining patients’ consent. This retrospective study consists of 144 OC patients classified using histopathological features mainly Tumor Node Metastasis (TNM) classification system. Using the patients’ medical records, all pathological and clinical data were collected following IRB approval (189–14).
2.2. Tissue microarray procedure
The FFPE tissue samples were transferred into a TMA format. From each block (donor block), sections were prepared and stained with hematoxylin and eosin to determine the tumor regions. From each block cylinders of tissues (0.6 mm in diameter) were punched from tumor areas and transferred into recipient block by a fully automated instrument to produce five TMA paraffin blocks used for the study. Sections of 4 mm thickness were cut from the resulting TMA block and mounted on glass slide. Human normal placenta tissue was used as a positive control with each run.
2.3. Automated immunostaining
Immunohistochemistry (IHC) was performed on OC tissues section slides using an automated staining system (Benchmark XT, Ventana Medical System, Inc. Tucson, Arizona, USA). Following the steps of deparaffinization, heat pretreatment then antigen retrieval. the anti-TIMP3 primary rabbit polyclonal antibody (TIMP3; Lot# ab93637; Abcam, Cambridge, UK) antibody (1:300 dilution) was then applied manually and incubated at room temperature for one hour. When the run was completed, the slides were rinsed by mild detergent followed by water to remove the residual buffer, then immersed into different alcohol buffers concentrations of (70%, 95% and 100%, respectively). The slides were washed in xylene solution repeatedly for three (3) minutes followed by two (2) minutes in each washing session. After that, one drop of DPX mounting medium was applied onto each TMA slide and covered by glass coverslip.
2.4. Scoring and evaluation of biomarkers expression
The evaluation of TIMP3 protein expression of all OC was assessed and scored manually by two independent expert molecular pathologists by using regular Nikon light microscope at ×40 magnification in a blind fashion to the clinicopathological parameters of patients. The intensity of staining and the fraction of positively stained cells were used to calculate the staining index score by the following formula:
Where (I) is the staining index and (f0 to f3) are the fractions of the cells showing a level of staining intensity (from 0 to +3) [Citation23].
The staining was thereafter categorized into two groups: (1) low (no/weak) expression and (2) high (moderate/strong) expression.
2.5. Statistical analysis
Statistical analyses were performed using the SPSS® (IMB NY, USA) software packages (PASW Statistics for Windows, version 19). Frequency tables were analyzed using the Chi-square test to assess the significance of the correlations between the categorical variables. The disease-specific survival (DSS) and disease-free survival (DFS) were calculated as the time from diagnosis to death (due to disease) or to the date of last seen alive, and time from diagnosis to the appearance of recurrent disease or date of last seen as disease free, respectively. In calculating DSS, patients died of other or unknown causes were censored. Univariate analysis for the survival outcomes such as DSS and DFS were based on Kaplan–Meier method, with log-rank (Mantel–Cox) comparison test. All tests with p-values <0.05 were considered as statistically significant.
3. Results
Our results showed that OC in Saudi population is more prevalent in younger females (age <50 years) than the older ones (age >50 years). Interestingly, most of our patients were obese (BMI > 26) which may increase the risk of OC ().
Table 1. Correlation between cytoplasmic TIMP3 protein expression patterns and clinico-pathological features of OC
3.1. TIMP3 protein profiling in ovarian cancer
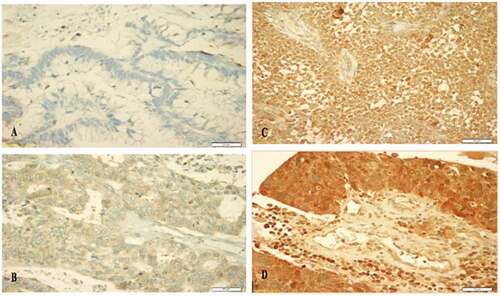
The expression of TIMP3 protein was mainly cytoplasmic (). The frequencies of the expression patterns of TIMP3 protein in 144 of OC samples evaluated by IHC technique revealed that: no expression (0) was observed in 2% of the analyzed cohort, weak expression (+1) in 16%, moderate expression (+2) in 33% and strong expression patterns (+3) in 49% of the cohort ().
3.2. Correlation of IHC TIMP3 protein expression with clinico-pathological features
Our data showed that age, tumor size and the involvement of lymph nodes had no significant associations with TIMP3 cytoplasmic protein expression patterns. However, a significant correlation of cytoplasmic TIMP3 protein expression patterns was observed with the tumor stage (p value = 0.05) and was borderline with the endpoint status (p value = 0.07) ().
3.3. Correlation Of IHC cytoplasmic TIMP3 protein expression with survival outcomes
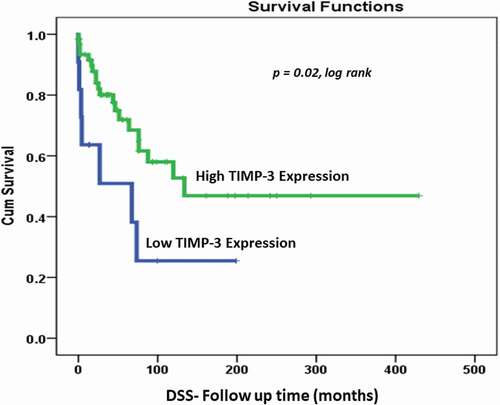
Kaplan–Meier survival analysis showed a significant correlation (p value = 0.02, log rank) between TIMP3 expression levels and DSS. In that, patients with high TIMP3 protein expression lived significantly longer (longer DSS) compared with those with lower TIMP3 expression (). At 10 years follow-up period, 78% of OC patients with lower expression of TIMP3 protein were dead compared to only 50% of those with higher TIMP3 expression, indicating a favorable prognosis for patients with higher MMP3 expression.
Figure 2. Cytoplasmic TIMP3 expression (0,1 vs 2,3) as determinant of disease specific survival (DSS) in univariate (Kaplan–Meier) analysis (p = 0.02, log rank)
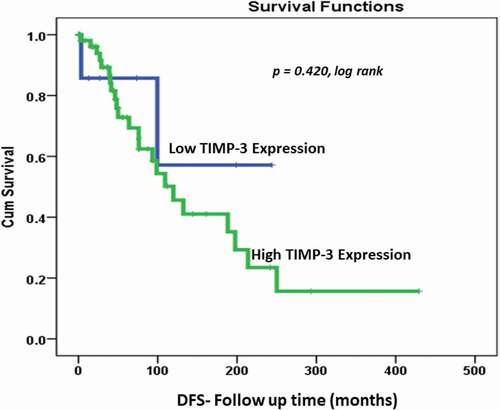
However, there was no significant correlation (p value = 0.42, log rank) of DFS between patients with high TIMP3 protein expression and living with no recurrence as compared with those with low TIMP3 expression ().
3.4. Cox’s regaression statistical analysis
Multivariate Cox’s regression analysis suggests that low TIMP3 protein expression pattern is an independent poor survival marker in relation to patient’s age, lymph node status, tumor grade, tumor stage and most importantly independent of the histological subtype of the tumor ().
Table 2. Cox’s regression analysis of the prognostic values of TIMP3 protein expression pattern, age at diagnosis, histological subtypes, lymph node status, tumor stage and grade
4. Discussion
OC remains the deadliest gynecological cancer due to the lack of early detection and/or effective treatments. Current OC management strategies based on clinical manifestations, such as age, tumor grade, lymph vascular invasion, lymph node involvement and tumor size are not enough to enhance recovery or minimize disease’s relapse. This context reflects the complexity of OC diseases and the multifactorial characteristics that made their management challenging. As a result, laudable efforts have been made by the scientific community to demystify the OC molecular complexity using several high-throughput technologies developed in the post-genomics era. Since biomarkers are indicators that may reflect the biological processes of health and disease, they are often associated with clinical manifestations and/or disease outcome. Consequently, the identification and evaluation of potential biomarker(s) involved in the OC onset, progression, metastasis and/or drug resistance is useful towards early diagnosis, prognosis and better OC management [Citation24,Citation25].
Several biomarkers have been investigated during the last two decades including members of the MMPs and TIMPs protein families. Studies showed that TIMPs are involved in several cancer biological processes including ECM maintenance and drug sensitivity [Citation26]. Moreover, TIMPs have also been studied for their potential role as prognostic biomarkers [Citation27] and therapeutic benefits for anticancer therapy [Citation28]. In fact, TIMPs are multifunctional proteins that inhibit the proteolytic activity of MMPs, thus suppressing metastasis [Citation29]. Members of TIMP family include TIMP-1, −2, −3 and TIMP-4. The TIMP3 (24-kDa glycoprotein) located at 22q12.3, has been shown to play a tumor suppressor role by inhibiting tumor angiogenesis, invasion, and metastasis. It is the only member of TIMP family that can bind to the extracellular matrix tightly and seems to be a key player in metastasis regulation [Citation30,Citation31]. Therefore, TIMP3 could be a key biomarker associated to OC metastasis, which is a dynamic complex multistep process involving cell detachment, migration, and invasion, EMT, disruption of tissue ECM movement, and establishment in distant location, cell proliferation and angiogenesis [Citation32].
In the current study, we studied the TIMP3 protein expression in OC samples and evaluated its potential role as OC prognosticator.
It is well documented that the risk of OC is strongly related to age, which means that OC is highly prevalent in older females compared to younger ones [Citation33]. Our results showed an early onset of OC in Saudi population (55% of cases aged less than <50 years). The Saudi Cancer Registry reported that most women diagnosed with OC were aged between 45 and 59 years old [Citation34] while in the UK, according to Cancer Research UK, over half (53%) of OC women were diagnosed at 65 or more [Citation35]. Therefore, additional investigative studies are needed to gain insights into the intricate molecular events that promote early onset of OC in the Arabic peninsula. Our results showed also that tumor size and the involvement of lymph nodes in OC had no significant relationship with the expression pattern of cytoplasmic TIMP3 protein. The endpoint status (survival vs death) correlated with TIMP3 expression in a borderline significant (p value = 0.07) manner, while the stage of tumor was significantly associated with cytoplasmic TIMP3 expression in OC (p value = 0.05). The increased expression of TIMP3 at advanced OC stages may be to counterbalance the enhanced proteolytic activity of MMPs and therefore slow down/prevent OC progression of the disease. In line with our findings, Kornfeld et al. in 2011 reported that in squamous cell carcinoma of the head and neck (HNSCC), TIMP3 mRNA expression levels did not associate with clinicopathological features in the tumor tissues, however there was a significant correlation between higher stages and increased TIMP3 mRNA expression (p value = 0.03) [Citation36]. Similarly, Su et al. reported that higher TIMP3 plasma levels in oral squamous cell carcinoma (OSCC) patients were significantly correlated with the tumor stage (p value = 0.009) and tumor invasion status (p value = 0.04) but not significant with the metastasis, lymph node status and cell differentiation [Citation37].
The Kaplan–Meier survival outcomes analysis of our OC cohort showed there was no significant correlation (p value = 0.42, log rank) in DFS between OC patients living with no recurrence despite a trend of higher recurrence in patients with higher cytoplasmic TIMP3 protein expression. However, we observed a significant correlation between high cytoplasmic TIMP3 protein expression in OC and DSS (p value = 0.02, log rank). This means that patients with higher cytoplasmic TIMP3 protein expression behave better and live longer than those with lower or negative cytoplasmic TIPM3. Multivariate Cox’s regression analysis suggests that low TIMP3 protein expression pattern is an independent poor survival marker in relation to other selected clinicopathological features. Patients with tumor of low TIMP3 have 20 times poor survival outcomes comparing to tumors with high TIMP3 expression pattern (p value = 0.025) . In line with these findings, Cymbaluk-Płoska et al. in 2018 showed that the high mean levels of TIMP3 proteins in OC patients’ serum correlate significantly with longer overall survival of patients [Citation38]. Recent findings by Huang et al. (2019) indicated that reduced levels of TIMP3 is associated with poor prognosis in colorectal carcinoma. In agreement with our data, the authors reported that patients with higher TIMP3 expression had longer recurrence periods. Also, Escalona et al., in 2018 proposed a model of TIMP-based therapy, they were suggesting that TIMPs (TIMP-1, -2, -3) may suppress the small residual tumors (chemo-resistant cells) after the chemotherapy which could increase the survival of OC patients and serve as therapeutic approaches [Citation19]. These results confirm our findings of a kind of TIMP3 protective (good prognosticator) effect against OC. TIMP3 unique properties and functions seems to be involved in several biological processes which involve promoter methylation of the TIMP3 gene, microRNA regulation and post-translational modification [Citation39–41]. It is even suggested to promote apoptosis in cervical and breast cancer [Citation42]. In addition, TIMP3 has also been reported to inhibit several cancer-related processes including cell migration, invasion and angiogenesis through direct interaction with endothelial vascular growth factor receptor-2 (VEGF-2) [Citation43–45].Our study showed that cytoplasmic TIMP3 protein expression could be used as a good prognosticator of OC patients mainly those at advanced OC. However, further investigations are required using additional larger and multi-institutional cohorts to gain in-depth understanding of the TIMP3-dependent molecular events involved in cancer progression and to investigate further the OC early onset phenomenon in Saudi patients.
Author Contributions
MA, AB and AC: Study design, contributed in statistical analysis, and manuscript drafting; MJ and TN: Contributed in data analysis and helped in results and discussions drafting; SH, HA, AA, MA and JA: Contributed in TMA design and construction, immunostaining and manuscript revision; KS: Contributed in clinicopathological data collection, data curation, and critical revision of the manuscript; MA: Supervision of the whole team. All authors critically reviewed and agreed on the final version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Oberaigner W, Minicozzi P, Bielska-Lasota M, et al. Survival for ovarian cancer in Europe: the across-country variation did not shrink in the past decade. Acta Oncol. 2012;51(4):441–7.
- Reid BM, Permuth JB, Sellers TA. Epidemiology of ovarian cancer: a review. Cancer Biol Med. 2017;14(1):9–32.
- Rossing MA, Daling JR, Weiss NS, et al. Ovarian tumors in a cohort of infertile women. N Engl J Med. 1994;331(12):771–776.
- Venn A, Watson L, Bruinsma F, et al. Risk of cancer after use of fertility drugs with in-vitro fertilisation. Lancet. 1999;354(9190):1586–1590.
- Dor J, Lerner-Geva L, Rabinovici J, et al. Cancer incidence in a cohort of infertile women who underwent in vitro fertilization. Fertil Steril. 2002;77(2):324–327.
- Cho KR, Shih I-M. Shih Ie M: ovarian cancer. Annu Rev Pathol. 2009;4(1):287–313.
- Sankaranarayanan R, Ferlay J. Worldwide burden of gynaecological cancer: the size of the problem. Best Pract Res Clin Obstetrics Gynaecol. 2006;20(2):207–225.
- Althubiti MA, Nour Eldein MM. Trends in the incidence and mortality of cancer in Saudi Arabia. Saudi Med J. 2018;39(12):1259–1262.
- Schorge JO, McCann C, Del Carmen MG. Surgical debulking of ovarian cancer: what difference does it make? Rev Obstet Gynecol. 2010;3(3):111–117.
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011 Jun 29;474(7353):609-15. doi: https://doi.org/10.1038/nature10166. Erratum in: Nature. 2012 Oct 11;490(7419):298. PMID: 21720365; PMCID: PMC3163504.
- Rankin EB. Genomics and molecular mechanisms of high grade serous ovarian cancer: the 12th biennial rivkin center ovarian cancer research symposium. Int J Gynecol Cancer. 2019;29(Suppl 2):s7–s11.
- Loret N, Denys H, Tummers P, et al. The role of epithelial-to-mesenchymal plasticity in ovarian cancer progression and therapy resistance. Cancers (Basel). 2019;11(6):838.
- Karam A, Dorigo O. MMPs in ovarian cancer as therapeutic targets. Anticancer Agents Med Chem. 2012;12(7):764–772.
- Birkedal-Hansen H, Moore WG, Bodden MK, et al. Matrix metalloproteinases: a review. Crit Rev Oral Biol Med. 1993;4(2):197–250.
- Schröpfer A, Kammerer U, Kapp M, et al. Expression pattern of matrix metalloproteinases in human gynecological cancer cell lines. BMC Cancer. 2010;10(1):553.
- Herszényi L, Hritz I, Lakatos G, et al. The behavior of matrix metalloproteinases and their inhibitors in colorectal cancer. Int J Mol Sci. 2012;13(10):13240–13263.
- Kleiner DE Jr., Stetler-Stevenson WG. Structural biochemistry and activation of matrix metalloproteases. Curr Opin Cell Biol. 1993;5(5):891–897.
- Murphy G, Knäuper V, Cowell S, et al. Evaluation of some newer matrix metalloproteinases. Ann N Y Acad Sci. 1999;878:25–39.
- Escalona RM, Chan E, Kannourakis G, et al. The many facets of metzincins and their endogenous inhibitors: perspectives on ovarian cancer progression. Int J Mol Sci. 2018;19(2):450.
- Rai GP, Baird SK. Tissue inhibitor of matrix metalloproteinase-3 has both anti-metastatic and anti-tumourigenic properties. Clin Exp Metastasis. 2020;37(1):69–76.
- Masson D, Rioux-Leclercq N, Fergelot P, et al. Loss of expression of TIMP3 in clear cell renal cell carcinoma. Eur J Cancer. 2010;46(8):1430–1437.
- Baird A, Esch F, Böhlen P, Ling N, Gospodarowicz D. Isolation and partial characterization of an endothelial cell growth factor from the bovine kidney: homology with basic fibroblast growth factor. Regul Pept. 1985;12(3):201–213.
- Buhmeida A, Dallol A, Merdad A, et al. High fibroblast growth factor 19 (FGF19) expression predicts worse prognosis in invasive ductal carcinoma of breast. Tumor Biology. 2014;35(3):2817–2824. .
- Kelloff GJ, Sigman CC, Scher HI. Biomarker development in the context of urologic cancers. Urol Oncol. 2015;33(6):295–301.
- Nedjadi T, Al-Maghrabi J, Assidi M, et al. Prognostic value of HER2 status in bladder transitional cell carcinoma revealed by both IHC and BDISH techniques. BMC Cancer. 2016;16(1):653.
- Brew K, Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim Biophys Acta. 2010;1803(1):55–71.
- Olivares-Urbano MA, Griñán-Lisón C, Zurita M, et al. Matrix metalloproteases and TIMPs as prognostic biomarkers in breast cancer patients treated with radiotherapy: a pilot study. J Cell Mol Med. 2020;24(1):139–148.
- Su CW, Lin CW, Yang WE, et al. TIMP-3 as a therapeutic target for cancer. Ther Adv Med Oncol. 2019;11:1758835919864247.
- Huang HL, Liu YM, Sung TY, et al. TIMP3 expression associates with prognosis in colorectal cancer and its novel arylsulfonamide inducer, MPT0B390, inhibits tumor growth, metastasis and angiogenesis. Theranostics. 2019;9(22):6676–6689.
- Gu X, Fu M, Ding Y, et al. TIMP-3 expression associates with malignant behaviors and predicts favorable survival in HCC. PloS One. 2014;9(8):e106161.
- Das AM, Koljenović S, Oude Ophuis CM, et al. Association of TIMP3 expression with vessel density, macrophage infiltration and prognosis in human malignant melanoma. Eur J Cancer. 2016;53:135–143.
- Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010;177(3):1053–1064.
- Tew WP. Ovarian cancer in the older woman. J Geriatr Oncol. 2016;7(5):354–361.
- Alghamdi IG, Hussain II, Alghamdi MS, et al. Incidence rate of ovarian cancer cases in Saudi Arabia: an observational descriptive epidemiological analysis of data from Saudi Cancer Registry 2001–2008. Int J Womens Health. 2014;6:639–645.
- Reid F, Bhatla N, Oza AM, et al. The world ovarian cancer coalition every woman study: identifying challenges and opportunities to improve survival and quality of life. International Journal of Gynecologic Cancer. 2021;31(2):238–244.
- Kornfeld JW, Meder S, Wohlberg M, et al. Overexpression of TACE and TIMP3 mRNA in head and neck cancer: association with tumour development and progression. Br J Cancer. 2011;104(1):138–145.
- Su CW, Su BF, Chiang WL, et al. Plasma levels of the tissue inhibitor matrix metalloproteinase-3 as a potential biomarker in oral cancer progression. Int J Med Sci. 2017;14(1):37–44.
- Cymbaluk-Płoska A, Chudecka-Głaz A, Pius-Sadowska E, et al. Suitability assessment of baseline concentration of MMP3, TIMP3, HE4 and CA125 in the serum of patients with ovarian cancer. J Ovarian Res. 2018;11(1):1.
- Yang H, Das P, Yu Y, et al. NDN is an imprinted tumor suppressor gene that is downregulated in ovarian cancers through genetic and epigenetic mechanisms. Oncotarget. 2016;7(3):3018–3032. .
- Petrovic N, Sami A, Martinovic J, et al. TIMP-3 mRNA expression levels positively correlates with levels of miR-21 in i n situ BC and negatively in PR positive invasive BC. Pathology - Research and Practice. 2017;213(10):1264–1270.
- Scilabra SD, Troeberg L, Yamamoto K, et al. Differential regulation of extracellular tissue inhibitor of metalloproteinases-3 levels by cell membrane-bound and shed low density lipoprotein receptor-related protein 1. J Biol Chem. 2013;288(1):332–342.
- Baker AH, George SJ, Zaltsman AB, et al. Inhibition of invasion and induction of apoptotic cell death of cancer cell lines by overexpression of TIMP-3. Br J Cancer. 1999;79(9–10):1347–1355.
- Sun JX, Zhang CX, Chen ZZ, et al. Cloning and expression of the cDNA encoding human tissue inhibitor of metalloproteinase-3 and its inhibition on angiogenesis. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao Acta Biochimica Et Biophysica Sinica. 1998;30(3):220–224.
- Zhang H, Wang YS, Han G, et al. TIMP-3 gene transfection suppresses invasive and metastatic capacity of human hepatocarcinoma cell line HCC-7721. HBPD INT. 2007;6(5):487–491.
- Han XG, Li Y, Mo HM, et al. TIMP3 regulates osteosarcoma cell migration, invasion, and chemotherapeutic resistances. Tumor Biology. 2016;37(7):8857–8867.