ABSTRACT
Introduction
Oral mycobiome profiling is important to understand host–pathogen interactions that occur in various diseases. Invasive fungal infections are particularly relevant for patients who have received chemotherapy and for those who have HIV infection. In addition, changes in fungal microbiota are associated with the worsening of chronic conditions like atopic dermatitis (AD). This work aims, through a systematic review, to analyze the methods used in previous studies to identify oral fungi and their most frequent species in patients with the following conditions: HIV infection, leukemia, and atopic dermatitis.
Methods
A literature search was performed on several different databases. Inclusion criteria were: written in English or Portuguese; published between September 2009 and September 2019; analyzed oral fungi of HIV-infected, leukemia, or AD patients.
Results
21 studies were included and the most identified species was Candida. The predominant methods of identification were morphological (13/21) and sugar fermentation and assimilation tests (11/21). Polymerase chain reaction (PCR) was the most used molecular method (8/21) followed by sequencing techniques (3/21).
Conclusions
Although morphological and biochemical tests are still used, they are associated with high-throughput sequencing techniques, due to their accuracy and time saving for profiling the predominant species in oral mycobiome.
Introduction
Fungi are eukaryotes that are dispersed across various ecosystems and terrestrial environments, totalizing a seven-digit number of species [Citation1]. However, estimates suggest that only 600 species are capable of causing disease in humans [Citation2]. Nevertheless, invasive fungal infections are still underestimated both in the field of research and procedural guidelines by health agencies around the world [Citation2]. Their epidemiology is still little understood due to the scarcity of studies, despite the fact that several fungal diseases such as malaria and tuberculosis are lethal [Citation2]. Around 1.6 million people die each year from fungal infections [Citation3], which are more challenging than infections by other microorganisms, in terms of the effectiveness of medicines available and time frame for administration [Citation4]. In addition, several anthropogenic factors contribute to the appearance of specific and resistant forms [Citation5], as evidenced by outbreaks of Candida auris in hospital settings [Citation4,Citation6].
The oral cavity is considered an important gateway for microorganisms (bacteria, fungi and viruses) that may be associated with localized oral problems such as periodontitis and candidiasis, or systemic diseases, including respiratory infections and cancer [Citation7]. The oral cavity mycobiome consists of somewhere around 100 fungal species, where each individual presents an average of 9 to 23 species and 75 different genera [Citation8]. Included among the most frequent genera are Candida, Cladosporium, Aureobasidium, Aspergillus and Malassezia spp., which are normally present in 25–75% of individuals [Citation9]. Oral fungal infections are usually caused by Candida species, especially C. albicans, which is found to be the most prevalent and pathogenic due to its adherence ability, to form germ tubes and produce extracellular proteases [Citation10–Citation13]. These microorganisms are of great importance to oral health, as they can stimulate the immune system in a very different way to bacteria. Furthermore, they have the ability to form filamentous hyphae, which provide a support system for the formation of biofilms between fungal and bacteria species [Citation14]. In the consortium of oral microbiota, in addition to fungal species, there are about 600 species of bacteria that are intrinsically associated with a person’s immune status. Dysbiosis is observed in several diseases such as infections by Candida spp. (oral candidiasis) and by Aspergillus fumigatus [Citation15]. The incidence and severity of these infections, as well as the causative pathogens, is closely related to several factors, such as immune status, geographic location, and underlying diseases [Citation16].
Accumulative evidence, since the first acquired immunodeficiency syndrome (AIDS) cases were reported in the 1980s, shows that patients infected with human immunodeficiency virus (HIV) are more susceptible to invasive fungal infections (IFI) [Citation14,Citation17]. HIV-infected patients present anti-fungal immune response failure, due to the effects of the virus in the lymphocytoid and myeloid cell lineages and the innate and adaptive response [Citation17]. HIV impairs the immune system by depleting CD4+ memory T-cells, and this reduces the effector T-cell population [Citation18]. HIV-infected patients are known to present lower Th2 cytokines levels than healthy subjects, and this is associated with Pneumocystis pneumonia [Citation19], and the impairment of tumor necrosis factor-α (TNF-α) and interferon-γ in response to cryptococcal meningitis [Citation17]. Despite their susceptibility to IFI caused by acquired fungi, several findings suggest that the HIV infection promotes a shift in fungal commensal microbiota that can lead to other infections. Oropharyngeal candidiasis (OPC) is the most common opportunistic disease found in the oral cavity and throat of HIV-infected patients, and it is strongly associated with CD4 + T-cells counts below 200 cells/mm3 [Citation17,Citation20,Citation21]. Various studies have demonstrated that OPC symptoms can be ameliorated by anti-retroviral therapy (ART), and this is associated with immune restoring by viral load reduction and regeneration of CD4 + T-cells [Citation20–Citation22]. Interestingly, the effect of ART on OPC does not seem to be related to Candida colonization but rather to the development of an inflammatory response towards microbial antigens due to immune reconstitution [Citation20,Citation23]. This suggests that oral fungal microbiota could be modulated in such patients both by the causative pathogen (HIV) and its treatment. OPC in HIV-infected individuals is not only caused by Candida albicans, but also other species from this genus such as Candida glabrata and Candida tropicalis, which have been isolated in various studies [Citation20].
IFI also represents a significative factor of morbidity and mortality for acute leukemia (AL) patients [Citation24]. The increased risk of IFI in these patients relies on a combination of several factors related to the disease itself, the status of the patients’ health and the treatment modality [Citation25]. In general, AL is characterized by an abnormal proliferation and differentiation of bone marrow progenitor cells that originate the myeloid and lymphoid cell lineages [Citation25]. Acute myeloid leukemia (AML) leads to neutropenia and impaired granulocyte functions, and immature myeloid cells can inhibit T-cell specific antigen response [Citation25]. On the other hand, acute lymphocytic leukemia (ALL) is characterized by the proliferation of poorly differentiated lymphoid cells [Citation26]. Neutrophil-depletion is believed to be the main factor that predicts a higher risk of IFI in AML than in ALL, with exception of Pneumocystis carinii pneumonia that is more common in the latter [Citation24,Citation25,Citation27]. Despite of these differences, AML and ALL treatments have high doses of cytotoxic drugs in common, which results in leukopenia [Citation25]. In addition, radio- and chemotherapy injure the mucosal barrier and this creates a propitious entrance for opportunistic pathogens [Citation25]. The most commonly isolated fungi from AL patients are Candida, Fusarium, Aspergillus, Tricosporum, and Pneumocystis [Citation25]. There is evidence that highly cytotoxic drugs also promote dysbiosis in commensal microbiota, as seen in the gut of lymphoma patients that received myeloablative treatment pre-hematopoietic stem cell transplantation [Citation28]. A recent longitudinal cohort study demonstrated that oral mycobiome is affected by remission-induction chemotherapy in AML patients, and that bacterial infection outcomes are related to modulations in Candida and Fusarium populations [Citation29].
Fungal dysbiosis is particularly relevant in chronic conditions other than HIV-infection and is also related to immune imbalance. Atopic dermatitis (AD) is an inflammatory skin condition in which patients present overreaction to harmless antigens in both topic and airborne forms (mainly in adults) and also allergy to some types of food (especially in children) [Citation30]. AD pathogenesis is believed to be mediated by deregulation of adaptive and innate responses associated with the disruption of the cutaneous barrier [Citation30]. AD patients present abnormal levels of Th2 cytokines that is observed by increase of IL-4 and IgE levels, causing an immediate hypersensitivity reaction [Citation31]. The progression of AD is related to fungi diversity. AD patients may have Malassezia on their skin and this can play a role in worsening AD patients’ conditions [Citation32]. Even though most AD symptoms manifest themselves in the skin, AD exacerbation and attenuation are also related to Candida in other body sites [Citation32]. Candida albicans was cultured from the nasopharynx of 59% of AD patients and its IgE-production stimulation was positively correlated to the severity of the disease [Citation33]. Although the major studies in atopic dermatitis concern fungal characterization in the skin, the oral cavity presents a similar architecture but is rarely investigated [Citation34,Citation35]. In a sectional study, AD patients presented greater colonization by Candida albicans in the mouth when compared to healthy controls [Citation36]. Interestingly, these patients presented lower serum levels of IgG against this species, which indicates alterations in the oral fungal microbiota [Citation36]. Fungal microbiota in AD can be affected by therapy modalities. Outpatients that present moderate and severe forms of AD are in general treated with immunomodulators and corticosteroids, the latter being related to oral candidiasis and favoring cariogenic activity [Citation34].
This body of evidence strongly suggests that fungal colonization, diversity, and interactions in oral microbiota are relevant for clinical outcomes of immune-mediated diseases, even though for those that have completely different etiologies. Moreover, these diseases often require hospitalization, which favors further colonization by opportunistic fungi [Citation33]. Nosocomial fungal infections are difficult to diagnose and the occurrence of dysbiosis in these patients can aggravate their clinical condition, therefore, the techniques used to identify these microorganisms must become faster and more accurate [Citation33,Citation36].
Despite the advances made in recent years, oral mycobiome is still a poorly understood subject. As a public health issue, mycoses are classically diagnosed through morphological analysis in cultures [Citation37,Citation38], and often the definitive conclusion is only possible in post-mortem exams [Citation24]. However, there are specific fungal stains that are effective for various species, such as Aspergillus spp., Candida spp., Coccidioides immitis, Blastomyces dermatitidis, and Sporothrix schenckii [Citation38]. Non-culture-based tests include immunodiffusion (ID), enzyme immunoassays (EIA) and the complement fixation test [Citation38]. Invasive aspergillosis can be detected by serum antigens such as galactomannan and 1,3-βD-glucan [Citation38]. One of the most reliable serological tests involves the detection of Cryptococcus neoformans antigens in cryptococcal meningitis [Citation38,Citation39]. However, in medical mycology the most promising diagnostic field is that of molecular tools [Citation39]. Methods involving polymerase chain reaction (PCR), mass spectrometry (MS) and sequencing are currently at the heart of the diagnostic revolution for invasive fungal infections [Citation4,Citation38].
The mycobiome field is underexploited and there is a bias due to the methods used for its characterization. Amplicon sequencing shows a lack of standardized methods by using different primers that can generate very distinguishable results [Citation40]. Usually, studies focus on analyzing mycobiome in a specific subject or when studying a bacteria-fungi consortium. Therefore, identification techniques must be improved so that fungal taxonomic classification can become more reliable [Citation41].
In order to understand the relevance of different mycobiome identification techniques, and their use in diseases that can react or perform differently in each individual, this systematic review selected three examples of diseases that are immune-mediated, present dysbiosis and are susceptible to nosocomial fungal infections: HIV-infection, leukemia, and atopic dermatitis. This study aims to identify the most prevalent fungal species in those diseases and the most affected sites. Additionally, this study aims to identify the more commonly used methods to identify the most prevalent fungi in the oral cavity of patients with these diseases.
Materials and methods
The present study is a systematic review. It was performed in agreement with the rules of PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) [Citation42] and was previously registered in PROSPERO under the protocol number CRD42020144972. In order to meet all the objectives proposed in this work, the following guiding questions were formulated for the preparation of this review: 1) What are the most common fungal species found in each disease? Is there a predominant species?/2) What were the sites of the oral cavity where the majority of the fungal species were isolated?/and 3) What are the most frequently used methods to identify fungi in the oral cavity in patients with one of these three diseases?
Search strategy
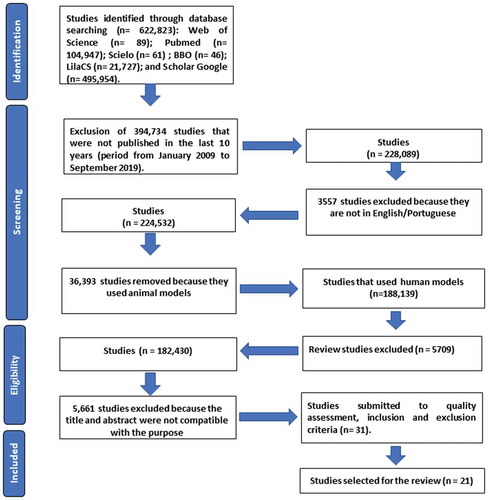
A flow chart (see ) was used to select the articles in this bibliographic survey in three online databases in the health field: MEDLINE, Web of Science and Latin American and Caribbean Health Sciences (LilaCS) and four portals were also included in the search: PubMed which belongs to MEDLINE, Scientific Electronic Library Online (SciELO), Google Scholar, and Brazilian Library of Dentistry (BBO). The search was performed using the Boolean operators (AND, OR and NOT) and combined with the keywords previously checked by MEDLINE MeSH described below: ‘fungi’, ‘mouth’, ‘microbiota’, ‘atopic dermatitis’, ‘HIV’, ‘mass spectrometry’, ‘leukemia’, ‘mycobiome’, ‘18S’. These words were selected because of their relationship to the topic of interest. In addition, the term ‘fungi identification’ was used despite not being described in MeSH. The inclusion criteria included, original articles published in the last 10 years (period from January 2009 to September 2019); works published in full in the following languages: Portuguese and/or English and research with compatible titles and abstracts related to the topic of interest. The exclusion criteria were: articles whose theme was not related to the objective of the study; articles that used a language other than those accepted by the inclusion criteria and those that used an animal model, as well as theses, books, dissertations, patents, and literature and/or systematic reviews. After finalizing the inclusion and exclusion criteria, the quality assessment was performed for further descriptive analysis of the selected studies.
Populations, Interventions, Control, Outcomes (PICOs)
We used the following PICOs criteria in our systematic review. Populations: leukemia, HIV-infected and atopic dermatitis patients who have been evaluated for the presence of fungal species in sites of their oral cavity. These patients may or may not have fungal lesion sites. Interventions: the present study will evaluate the methods used to identify fungal species in HIV-infected, atopic dermatitis and leukemia patients and thus verify the efficiency of each one. Control: healthy individuals with no fungal lesions in the oral cavity. Outcomes: 1) Identify and detail the main fungal profiling methods used for HIV, leukemia, and atopic dermatitis patients. 2) Identify the predominant fungal species found in HIV, leukemia, and atopic dermatitis patients. 3) Compare oral mycobiome between HIV, leukemia, and atopic dermatitis patients with control patients. 4) Identify the oral sites of patients with atopic dermatitis, leukemia, and HIV in which there is a predominance of isolated fungal species.
Study selection and quality assessment
Two independent reviewers selected the studies and cross checked the keywords previously checked by MEDLINE MeSH. After, these two reviewers analyzed the titles and abstracts of each study independently. Duplicates were excluded and studies that were selected had their full-text analyzed by the two reviewers. If any disagreement occurred, a third reviewer was consulted. Data were summarized and the studies that met the inclusion and exclusion criteria had their methodology analyzed through quality assessment () by the two reviewers. The quality assessment was based on the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) criteria and established by the two reviewers (C.S.S and P.M.G.R.). Each item of the quality assessment presented a score of 1 point. Studies were classified as level A (high methodological quality) if they received scores between 7 and 9 indicates. If they scored between 5 and 6 points were classified as level B (intermediate methodological quality). Studies that were classified as level A and B were included in this study, and if they scored below or equal to 4 points (level C- low methodological quality) were excluded.
Table 1. Quality assessment of the selected studies for the systematic review.
Results
In the initial search (), a total of 622,823 studies were found in 2 portals and 4 databases: Web of Science (n = 89); PubMed (n = 104,947); SciELO (n = 61); BBO (n = 46); LilaCS (n = 21,727); and Google Scholar (n = 495,954). After the inclusion and exclusion criteria were applied, there were a total of 49 studies, and after their duplicates had been removed, there were 31 articles left. Then, the quality assessment was carried out, which studies that were classified with high and intermediate methodological quality were selected. After the quality assessment, 21 studies were selected for this systematic review ().
Characteristics of selected articles
Among the 21 studies, 11 defined their type of study as: cohort (6/11); prospective (3/11); and transversal (2/11). All studies were in English and showed a great variability in the number of samples, ranging from 24 to 990 patients. Most of the studies addressed HIV-infection, with 18 studies in total, while atopic dermatitis (1/21) and leukemia (2/21) were minorities. Also, most of the studies used adult patients (16/21), however, five articles evaluated pediatric patients: three studies involving HIV-infected patients and the two studies that evaluated leukemia patients. Another aspect observed was the method to collect the clinical specimens: the oral swab (9/21) and the oral rinse (6/21) were the most commonly used methods to collect from the oral cavity. The oral rinse can evaluate not only the Candida load but also detect other microorganisms, while the oral swab is more suitable for specific sites of microbiological examination [Citation43].
The identification of the species showed that most studies identified Candida species, especially C. albicans (12/21), C. tropicalis (13/21), C. parapsilosis (12/21) and C. dubliniensis (11/21). Articles that identified fungi in HIV patients also found fungi genera other than Candida, such as: Malassezia, Debaryomyces, and Aspergillus [Citation44–Citation47]. The description of the selected studies is given in . The species identified in each study and their main objectives are described in .
Table 2. Description of studies selected by the quality assessment.
Table 3. Isolated species and study objectives.
Fungal colonization in HIV-infected patients
The oral mycobiome of HIV-infected individuals has been demonstrated to be different when compared with non-HIV-infected patients [Citation48], and is most likely to be correlated with the immunodeficiency observed in HIV-infected individuals [Citation49]. Distinct microbial communities in the digestive tract of HIV-infected individuals under antiretroviral therapy (ART) can be found; furthermore, patients with a history of AIDS, displaying T CD4+ lymphocytes count < 200 cells/mm3, have significantly lower subgingival plaque fungal diversity [Citation49]. The interaction in both gut and oral bacterial diversity can be correlated with the use of specific ART regimens. For instance, the use of protease inhibitors (PI) with nucleoside reverse transcription inhibitors (NRTI) can be associated with decreased oral diversity compared to an NRTI + non-nucleoside reverse transcription inhibitors (NNRTI) regimen. In the gut, on the other hand, integrase strand transfer inhibitors (INSTI) combined with NRTIs can demonstrate significantly higher diversity compared to a combination of NRTI and NNRTI [Citation49]. These imply long-term effects of HIV-infection on the mycobiome as well as the bacterial microbiome, which is either not completely mitigated by ART or represents effects of the treatment itself with disruptions of the gut barrier integrity [Citation49].
The results of this review point to the importance of the genus Candida in the oral colonization of patients with HIV. C. albicans was present in 11 HIV studies selected in our review, as well as C. tropicalis which was found in 13 studies [Citation44,Citation47,Citation50–Citation60]. Although the presence of Candida spp. in the most of the articles that focused on HIV-infected patients, Mukherjee et al. [Citation44], Mukherjee et al. [Citation53] and Fukui et al. [Citation45] through sequencing identified genera other than Candida. Mukherjee et al. [Citation44] found differences between the HIV-infected mycobiome and uninfected individuals. The authors found a prevalence of Candida, Epicoccum and Alternaria in HIV-infected patients (92, 33, and 25%, respectively), while in uninfected individuals the most frequent genera were Candida, Pichia and Fusarium (58%, 33% and 33% respectively). The core of the oral mycobiome in both HIV-infected and uninfected individuals consisted of 5 genera of which only 2 (Candida and Penicillium) were common to both groups [Citation44]. Mukherjee et al. [Citation44] evaluated the correlation between different fungi that were presented in the mycobiome of HIV-infected patients and found 6 fungal–fungal interactions (Candida-Epicoccuum, Candida-Trichosporon, Epicoccum-Trichosporon, Penicillium-Corynespora, Penicillium-Fusarium, and Alternaria-Serpula) that were statistically significant (p≤ 0.03) [Citation44]. Mukherjee et al. [Citation53] analyzed the mycobiome of HIV-infected smoker and non-smoker patients using sequencing and bioinformatic analysis [Citation53]. The Shannon diversity index pointed to a significantly low diversity in the mycobiome of HIV-infected non-smokers compared to HIV-infected smokers and uninfected smokers but not at a genus level (p≤ 0.02) [Citation53]. The core mycobiome included 11 fungal genera in common in the three groups and 4 which were only detected in the HIV-infected smoker group (Cladosporium, Nakaseomyces, Scleroderma, and Rhodotorula) [Citation53]. At species level, 2 were unique to the HIV-infected smoker group (C. glabrata and Scleroderma spp.). Levels of Candida were higher in the HIV-infected and uninfected smoker groups (34.7% and 38.45, respectively) compared to HIV-infected non-smoker group (19.2%). In summary, the fungal diversity of the mycobiome was significantly greater in HIV-infected smokers compared to HIV-infected non-smokers [Citation53].
Fukui et al. [Citation45] used amplicon sequencing to compare HIV-infected and non-infected individuals. There was no significant difference regarding the mycobiome of the palatine tonsils between HIV-infected and non-infected individuals. At the phylum level, Ascomycota and Basidiomycota were abundant but no significant difference was observed and at species level Candida and Malassezia colonized both groups [Citation45].
Fungal colonization in patients with leukemia
In the present study, only 2 articles that related to fungal identification in patients with leukemia were selected. De Mendonça et al. [Citation61] and De Mendonça et al. [Citation62] conducted their studies in pediatric patients with acute lymphoid leukemia (ALL) who were undergoing chemotherapy during collection. Both studies were published by the same study group. Their methodology included material collection on the 14th day after the start of chemotherapy, a severity assessment of mucositis in the patients and microbiological analyses.
In the second study, De Mendonça et al. [Citation61] collected samples using mucosal swabs and found a significant association in their patients between the severity of mucositis with three variables: presence of herpes simplex virus type I (p = 0.0347), presence of Candida spp. (p = 0.0078) and low platelet count (p = 0.0064). While, in the first study De Mendonça et al. [Citation61] collected samples via an oral rinse and also observed the same significant association between the severity of mucositis with the presence of the herpes simplex virus type I (p = 0.0347) and the presence of Candida spp. (p = 0.0078), but did not include hematological factors in their work. De Mendonça et al. [Citation62] found that on the 14th day 25.4% of their patients were colonized by Candida. This value decreased to 14.9% of the patients on the 56th day. Although the authors did not refer to the possible causes of the observed reduction, they made it clear in their methodology that all patients received routine dental treatment during the study and that no antifungal agents were administered. In the second work of the group, there was a small reduction in the presence of Candida on the 56th day. The number of patients with severe mucositis appeared to be greater on the 14th day than on the 56th day.
Fungal colonization in patients with atopic dermatitis
The colonization of the cutaneous microbiota of patients with AD is different from healthy individuals, with Staphylococcus aureus being the most commonly found microorganism [Citation63–Citation66]. This is propitiated due to defects in the function of the skin barrier and deficiency of ceramides that facilitate the penetration of microorganisms and allergens [Citation63–Citation66]. In atopic dermatitis, most studies are focused on the cutaneous microbiota, and few have verified the microbial oral status of these patients. Generally, the cutaneous mycobiome of patients with AD have Malassezia spp [Citation32,Citation67] and Candida spp [Citation36].
After our selection and quality assessment, only one article was included in this survey. The results verified the colonization of Candida and the humoral immune response specifically against the species C. albicans in the skin and oral cavity [Citation36]. This study found a higher frequency of colonization by C. albicans in patients than in controls (23% vs. 5%, p < 0.05), although there was no statistically significant difference in relation to colonization by the genus Candida. In general, the most isolated species were C. albicans and C. glabrata, both in the control group and in patients with AD. In another study reporting on the levels of immunoglobulins, Javad et al. [Citation36] found that 14% of AD patients colonized by Candida had IgM levels below 10 U/ml, while specific IgG levels were significantly lower for C. albicans in patients than in controls [Citation36]. This result may be linked to the fact that IgG plays a key role in the humoral immune response and may also be related to increased colonization of C. albicans in patients with AD [Citation36,Citation68].
Fungal identification methods
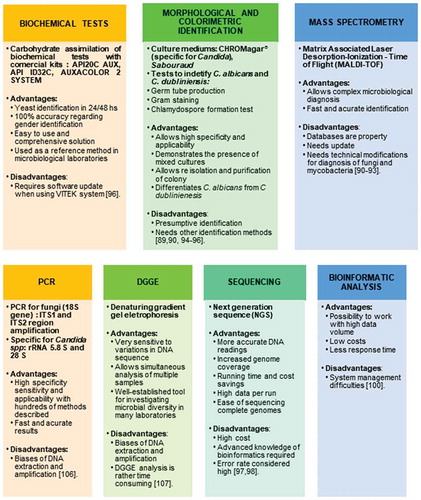
This study describes different methods to identify fungi (). Some are only presumptive, such as the colorimetric and morphological identification methods, and these were the most used technique in the selected articles (13/21), followed by sugar fermentation and assimilation tests (11/21), which are used specially to distinguish the different Candida species [Citation69]. Among the molecular methods, the polymerase chain reaction (PCR) was the most used (8/21) in the selected articles. This result demonstrates an evolution compared to the previous identification methods linked to microscopic aspects such as: the germ tube method (9/21) and specific means for the production of chlamydospore (8/21), which are tests used as a standard to identify the species of Candida albicans [Citation70]. Some studies did not use molecular techniques, but used biochemical tests to confirm the presumptive results of colorimetric and microscopic identification methods. Among these techniques, the most widely used are the sugar fermentation and assimilation tests, which currently have several commercial kits that have facilitated and disseminated their use in laboratories such as API20 C AUX (Biomérieux®), which is one of the most used commercial systems for the identification of yeasts [Citation71]. The articles that used commercial kits to identify the most common Candida species were: Das Chagas et al. [Citation50]; Junqueira et al. [Citation51]; Mane et al. [Citation70]; Merestein et al. [Citation55]; Li et al. [Citation47]; Jiang et al. [Citation57]; Thanyasrisung et al. [Citation58]; Blignaut and van Heerden [Citation72]; Menezes et al. [Citation59]; Lourenço et al. [Citation60]; and Vijendran et al. [Citation46]. Mushi et al. [Citation73] was the only work that used MS focused on microbiology to identify fungi. They chose to perform MALDI-TOF spectrometry (‘Matrix Associated Laser Desorption-Ionization – Time of Flight’), which compares a mass spectrum of a microorganism that has been isolated with a spectrum of strains already known that are being used as references in the program library [Citation62,Citation63]. Blignaut and van Heerden [Citation72] also evaluated the genetic subtype of C. albicans from two different oral sites (tongue swab and dentin scrape) by using the denaturing gradient gel electrophoresis technique, better known by the acronym DGGE after having identified the species by the commercial kit ID32 C (Biomérieux®).
Figure 2. Most frequent methods used to identify fungi. Mycobiome identification can be divided into 7 general approaches, with each one presenting advantages and disadvantages.
The sequencing method was performed on three articles that evaluated HIV-infected patients [Citation44,Citation45,Citation53] and later Fukui et al. [Citation45] and Mukherjee et al. [Citation53] did the analysis by bioinformatics. Mukherjee et al. [Citation44] carried out pyrosequencing to analyze the microbiome and the characterization of nucleic acids, while Mukherjee et al. [Citation53] used the Ion Torrent sequencing platform, which uses pH to identify the bases [Citation74]. Fukui et al. [Citation45] used the Illumina platform which, like the Sanger method, performs sequencing by synthesis of the DNA polymerase enzyme and terminator nucleotides labeled with different fluorophores [Citation75].
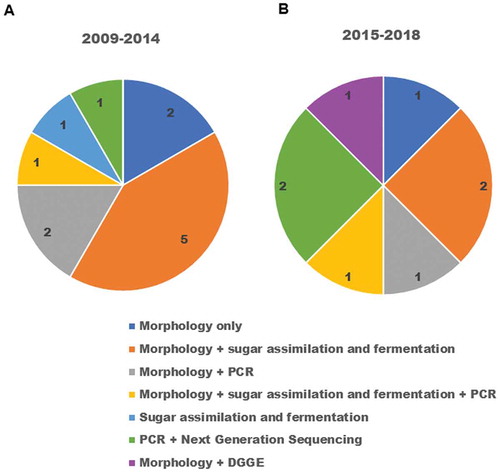
Over the years, the identification protocols have changed. Morphological methods are frequently being executed first and then the molecular approaches are applied next. Furthermore, this trend is not an exclusive feature of oral fungal identification but is a trend in the entire field of microbiology and its subset mycology [Citation8,Citation76–Citation79]. ) shows that the morphological methods were the most used in the period from 2009 to 2014 and these were followed by the sugar fermentation and assimilation tests in the same period. However, in the period from 2015 to 2018 ()) we can see that there is a balance between the morphological methods together with the sugar fermentation and assimilation tests and PCR and next-generation sequencing, which indicates the evolution of resources and diffusion of molecular techniques among research laboratories.
Discussion
The role of fungi as opportunistic pathogens has gained prominence in recent decades, mainly due to the emergence of the AIDS pandemic in the 1980s [Citation2]. Fungal infections are probably one of the main challenges for medicine in the 21st century, as they have been neglected in recent decades compared to other pathogens, such as bacteria and protozoa [Citation4]. Currently, there is a clear association between immune-mediated diseases and susceptibility to invasive fungal infections [Citation80,Citation81], which can lead to morbidity and clinical worsening of a patient’s health [Citation82–Citation84].The use of low cost and optimized methods to identify the genus and species of microorganisms in hospital environments are required so that they can be detected quickly and preventive measures can be taken against infection [Citation82–Citation84]. Conventional laboratory techniques using a microscope and culture medium require days until the microorganism is in the optimum growth phase and can be identified [Citation85,Citation86]. In addition, these techniques generate a high cost and the need for a team trained in a microbiology laboratory [Citation85]. An alternative technique that can be used routinely for fungi diagnosis is MS with matrix-assisted laser ionization and desorption source and time-of-flight analyzer (MALDI-TOF), which performs the diagnosis in a short time and precisely [Citation87–Citation89]. Currently, this technique also performs the identification of filamentous fungi after undergoing adaptations in the extraction process to obtain protein profiles [Citation89]. The use of morphological techniques for the identification of fungi is important to understand the evolution of morphological characteristics of fungi and to classify them according to their family and genus level [Citation90,Citation91]. However, this method is not very effective in identifying the species level, and is a challenge when performed by non-specialists [Citation90,Citation92]. Culture media are an example of morphological techniques that can be used to identify and distinguish species [Citation93]. This study demonstrated that despite presenting limitations, morphological techniques such as colorimetric identification CHROMagar® as well as the germ tube test are still widely used prior to the molecular techniques [Citation36,Citation52,Citation56,Citation72]. In this case, molecular techniques were used after these morphological analyses in order to confirm the species identified at the molecular level because the mycological identification of species becomes necessary when the investigated patient is already immunocompromised [Citation68].
In the present review, only the works of Mukherjee et al. [Citation44], Mukherjee et al. [Citation53], Fukui et al. [Citation45], Li et al. [Citation47] and Vijendran et al. [Citation46] identified genera other than Candida in HIV-infected patients. This can be explained due to the techniques used by their studies: Mukherjee et al. [Citation44] used the Multitag 454 Pyrosequencing; Mukherjee et al. [Citation53] used the Ion-Torrent sequencing platform; and Fukui et al. [Citation45] used the Illumina Miseq. Next-generation sequencing provides the identification and typing applications of all pathogens in a single protocol, which contrasts with Sanger sequencing [Citation93]. This advantage is of great use in medical microbiology and also for preventive measures against infection [Citation93]. However, next-generation sequencing requires basic knowledge in bioinformatics, which makes its use more difficult [Citation94]. Thus, when comparing the advantages and differences between sequencing techniques, we can see that the Ion Torrent platform, used by Mukherjee et al. [Citation53] is the simplest and also has a low cost, and consequently, it is widely available, especially in Europe and the USA [Citation74]. The Multitag 454 Pyrosequencing that was used in the work of Mukherjee et al. [Citation53] is more efficient than the Sanger method presenting greater sequence coverage capacity, precision, flexibility, parallel processing, easy automation potential, and instantaneous sequencing response [Citation95]. However, its use requires the use of numerous reagents, in addition to presenting a high cost and an error rate that is considered high (0.0098) [Citation96].
In relation to the sequencing platforms, there were some differences between the results, which may be linked to the platform used, as well as the distinct sample types. Fukui et al. [Citation45] used palatine tonsil swabs while the other works used oral rinses. The Illumina sequencing platform was the most used platform in the selected studies that investigated fungi in HIV-patients, however, it has a higher cost and a long analysis time [Citation80]. Fukui et al [Citation45] showed, with the use of the Illumina platform that the fungal diversity and its composition in the HIV-infected and non-infected groups were the same [Citation45]. In this work, the authors verified that the alpha and beta diversity of the mycobiome of the palatine tonsil between HIV-infected and non-HIV-infected patients did not have a significant difference. Unlike Fukui et al. [Citation45], the work of Mukherjee et al. [Citation53] found that the diversity of mycobiome in non-smoker HIV-patients was significantly less when compared to the diversity in patients with and without HIV but who were smokers [Citation50]. Regarding genus, Fukui et al. [Citation45] observed that Candida and Malassezia were the ones that most colonized both groups, with Penicillium being the significantly higher gender in the group without HIV [Citation45]. This result contrasts with the result of Mukherjee et al. [Citation44] in which there is a difference in oral mycobiome between HIV-infected and non-infected individuals. In infected individuals, the genus Candida was the most prevalent, affecting 92% of this group [Citation44]. Besides that, Mukherjee et al. [Citation44] found that a decrease in Pichia is related to an increase in Candida [Citation44]. Thus, the results of this study show us that there are changes in oral mycobiome associated with HIV [Citation44]. Mukherjee et al. [Citation53] unlike the other two studies that involved mycobiome sequencing and analysis in patients with and without HIV, compared HIV-infected individuals who were smokers and non-smokers to uninfected smokers. Consequently, they presented different results and verified high levels of Candida in individuals with and without HIV who smoked when compared to the non-smoking HIV group [Citation53]. Although the authors of the study did not hypothesize the cause of this reduction, several previous studies have shown that smoking is associated with greater colonization by Candida [Citation97–Citation101]. Li et al. [Citation47] and Vijendran et al. [Citation46] did not use next-generation sequencing techniques and identified several fungi in their studies besides Candida spp. While Vijendran et al. [Citation46] identified species of Candida and filamentous fungi such as Dermatophyte, Pityrosporum, and non-dermatophyte filamentous fungi; Li et al. [Citation47] found C. albicans, C. glabrata, C. parapsilosis, C. krusei, C. tropicalis, C. rugosa, C. norvegensis, C. lusitaniae, C. guilliermondii, Pichia ohmeri, and Saccharomyces cerevisiae. Li et al. [Citation47] performed colorimetric and morphological identification for Candida species using CHROMagar® and then used fermentation and assimilation tests through the API 20 C AUX system (bioMérieux, France). In addition, this study also used the chlamydospore production test to differentiate C. albicans and C dubliniensis that in the selective environment have a similar morphology and coloring [Citation47,Citation92].
Countless techniques are still used as a standard in the identification of fungi among the microscopic aspects. Tests of selective medium, germ tube test, hyphae production and specific medium for the production of chlamydospores are examples of morphological methods. De Mendonça et al. [Citation62], Dos Santos Abrantes et al. [Citation54] and De Mendonça et al. [Citation61] were the three works that only used morphological identification methods. De Mendonça et al. [Citation62] and De Mendonça et al. [Citation61] performed microbiological culture tests to identify Candida species present in ALL patients, but did not distinguish between species. The focus of Dos Santos Abrantes et al. [Citation54] was to identify Candida species present in HIV-infected patients with oral candidiasis so they used methods for morphological identification through the chromogenic medium followed by gram stain and germ tube test in order to distinguish C. albicans from C. dubliniensis.
The study by Mushi et al. [Citation73] was the only one that used MS with matrix-assisted ionization and a laser desorption source and time-of-flight analyzer (MALDI-TOF) in the identification and confirmation of non-albicans Candida species in HIV patients. This technique facilitates identification when compared to conventional methods because it is faster and has high accuracy for a great variability of microorganisms [Citation102,Citation103]. In addition to microscopic morphological and spectrometric techniques, we can also see that biochemical tests, more precisely the fermentation and assimilation tests, were also widely used in the HIV-infected articles selected in this systematic review (11/21). In 7 articles that studied HIV-infected patients, this test was performed after microbiological culture and morphological identification in order to confirm with greater accuracy the species found [Citation46,Citation47,Citation50,Citation51,Citation55,Citation57,Citation60].
Menezes et al. [Citation59], in addition to using morphological analyses, subsequently performed PCR to distinguish C. albicans from C. dubliniensis [Citation59], while Thanyasrisung et al. [Citation58] only used PCR for ambiguous results from their biochemical tests [Citation58]. Menezes et al. [Citation59] found 11 different species of Candida and nine associations between species of Candida, which made it stand out among the others due to the combination of three different identification techniques [Citation59].
Only Mane et al. [Citation70] that studied HIV-infected patients made use of fermentation and assimilation tests using API 20 C AUX (BioMerieux) without any other method of complementary identification [Citation70]. These tests have an accuracy of 92% on average, greater reliability in genus identification and greater discriminatory power when associated with a zymogram [Citation92]. Fermentation and assimilation tests are effective to identify yeasts since they evaluate the ability of a yeast to ferment a certain carbohydrate through the production of carbon dioxide [Citation92].
Among the other molecular techniques, PCR was very present in the identification of species in the selected articles, because of its high degree of sensitivity and selectivity [Citation98, Citation104]. This technique was used in eight articles, where seven of them analyzed HIV-infected patients [Citation36,Citation44,Citation45,Citation53,Citation56,Citation58,Citation59]. Sharifzadeh et al. [Citation56] and Menezes et al. [Citation59] used specific primers for C. albicans and C. dubliniensis species, while Moris et al. [Citation52] used other primers for C. parapsilosis.
In general, universal primers and amplicon sequencing produce a wide spectra of microbiota. To investigate mycobiota the internal transcribed spacer (ITS) between 18S and 28 S rRna genes is the most used locus [Citation83]. Mukherjee et al. [Citation44], Fukui et al. [Citation45] and Mukherjee et al. [Citation53] amplified the ITS region to detect the presence of several fungi. Javad et al. [Citation36] was the only study that amplified the D1/D2 domain, which identifies yeast species. Thanyasrisung et al. [Citation58] performed PCR using specific primers for five different Candida species. This shows that there is a diversity of choice in relation to the region to be amplified by PCR, which makes the study direct its objective, and obtain results faster.
Blignaut and van Heerden [Citation72] was the only work that made use of DGGE which is an excellent tool to assess previously unknown microbial diversity [Citation105]. Using this technique, the authors were able to see that colonization of oral soft tissue and dentin is done by the same genetic subgroup of C. albicans [Citation81]. DGGE presents fast and easy analytical methodologies, in addition to separating DNA fragments of the same size, but with different nucleotide sequences [Citation99–Citation101]. The technique has numerous advantages, such as: accessibility in poorly equipped laboratories, screening before sequencing, comparing the efficacy and reproducibility of different DNA extraction protocols and facilitating the interpretation of its results [Citation106–Citation108].
In chronic diseases like AD, C. albicans has been shown to be related to the disease, since the production of specific antibodies for C. albicans, is associated with the severity of AD, as in the study of Matsumura et al. [Citation105], where IgE antibody levels were significantly higher in patients with AD than in the control group [Citation105]. Javad et al. [Citation36] evaluated antibodies other than IgE such as IgG, IgA and IgM. The authors found statistically significant low levels of IgG for C. albicans in AD patients when compared to controls. IgG is an important factor in the humoral immune response and is one of the most important serum immunoglobulins [Citation98]. Thus, we can emphasize that the genus Candida is one of the main yeasts that can lead these patients to acquire infections such as oral candidiasis, which may be systematically disseminated by mucositis lesions or when it reaches the oropharynx [Citation19,Citation109].
Conclusion
Knowledge of the most frequent fungal species in AD, leukemia and HIV is important in order to contribute to the improvement of the clinical condition of the disease, as well as an alert of the patient’s general condition of health. Patients in hospital are at greater risk of acquiring fungal infections, especially those who are immunosuppressed or in critical condition. Therefore, the characterization and monitoring of fungi in hospital environments, of the fungal microbiota on the hands of health professionals and colonized sites of patients are necessary measures to control these microorganisms.
In this systematic review, we found that the genus Candida was the most identified among the three diseases studied, with C. albicans, C. parapsilosis, and C. tropicalis being the species most frequently found. The most frequently identified colonized sites in the selected studies were the oral mucosa and dorsal surface of the tongue. In the field of diagnosis, there is a great diversity of methods used to identify fungi in medical microbiology. However, many of them are still unviable, especially in terms of cost, training of personnel and speed of diagnosis. Despite presenting low precision and only presumptive identification of species, morphological identification by microscopic tests is still widely used in the identification of fungi of patients with leukemia, HIV, and AD. This technique was used before the introduction of biochemical and/or molecular tests that, by providing greater precision, are able to identify species with greater predictability and fewer errors. However, nowadays next-generation sequencing is gradually coming available for these identification tasks and can provide an accurate molecular diagnosis, as well as the quantification of the predominant species in medical microbiology. However, it is not to be seen as a breakthrough for oral fungal identification but rather as a reflection of a shift in the whole field of microbial surveillance.
Author contributions
C.S.S. and D.C.F. conceived the study. C.S.S. and P.M.G.R. performed the literature search, the data extraction and analysis. C.S.S., P.M.G.R. and M.S.V. wrote the draft manuscript. M.S.V and A.M.P.S. performed a critical appraisal. L.S.G, M.G.R. and D.C.F. revised and approved the final version of the manuscript. All the authors agreed with the submission for publication.
Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Funding
References
- Bass D, Richards TA. Three reasons to re-evaluate fungal diversity on earth and in the ocean. Fungal Biol Rev. 2011;25:159–19.
- Brown GD, Denning DW, Levitz SM. Tackling human fungal infections. Science. 2012;336:647.
- Anonymous. Stop neglecting fungi. Nat Microbiol. 2017;17120. DOI:10.1038/nmicrobiol.2017.120
- Casadevall A. Fungal diseases in the 21st century: the near and far horizons. Pathog Immun. 2019;3:183–196.
- Gostincar C, Grube M, Gunde - Cimerman N. Evolution of fungal pathogens in domestic environments? Fungal Biol. 2011;115:1008–1018.
- Garcia-Solache MA, Casadevall A. Global warming will bring new fungal diseases for mammals. mBio. 2010;1(1):e00061–10.
- Chandra J, Retuerto M, Mukherjee PK, et al. The fungal biome of the oral cavity. Methods Mol Biol. 2016;1356:107–135.
- Ghannoum MA, Jurevic RJ, Mukherjee PK, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PloS Pathog. 2010;6(1):e1000713.
- Dupuy AK, David MS, Li L, et al. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One. 2014;9(3):e90899.
- Neville BW, Damm DD, Allen CM, et al. Oral & Maxillofacial Pathology. Missouri: Elsevier; 2016.
- Sherman RG, Prusinski L, Ravenel MC, et al. Oral Candidoses. Quintessence Int. 2002;33:521–532.
- Simões RJ, Fonseca P, Figueira MH. Infecções por Candida spp na Cavidade Oral. Odontol Clín Cient. 2013;12:19–22.
- Budtz-Jorgensen E. Etiology, pathogenesis, therapy and prophylaxis of oral yeast infections. Acta Odontol Scand. 1990;48:61–69.
- Cui L, Morris A, Ghedin E. The human mycobiome in health and disease. Genome Med. 2013;5:63.
- Peters BA, Wu J, Hayes RB, et al. The oral fungal mycobiome: characteristics and relation to periodontitis in a pilot study. BMC Microbiol. 2017;17:157.
- Binder U, Lass-Flörl C. Epidemiology of invasive fungal infections in the mediterranean area. Mediterr J Hematol Infect Dis. 2011;3. DOI:10.4084/MJHID.2011.0016.
- Armstrong-James D, Meintjes G, Brown GD. A neglected epidemic: fungal infections in HIV/AIDS. Trends Microbiol. 2014;22:120–127.
- Okoye AA, Picker LJ. CD4(+) T-cell depletion in HIV infection: mechanisms of immunological failure. Immunol Rev. 2013;254(1):54–64.
- Myers RC. STAT4-Dependent and -Independent Th2 Responses Correlate with Protective Immunity against Lung Infection with Pneumocystis murina. Immunol. 2013;190:6287–6294.
- Patil S, Majumdar B, Sarode SC, et al. Oropharyngeal Candidosis in HIV-Infected Patients-An Update. Front Microbiol. 2018;9:980.
- Tamí-Maury IM, Willig JH, Jolly PE, et al. Prevalence, incidence, and recurrence of oral lesions among HIV-infected patients on HAART in Alabama: a two-year longitudinal study. South Med J. 2011;104(8):561–566.
- Thompson MA, Aberg JA, Cahn P, et al. Antiretroviral treatment of adult HIV infection: 2010 recommendations of the International AIDS Society-USA panel. JAMA. 2010;304(3):321–333.
- Walker NF, Scriven J, Meintjes G, et al. Immune reconstitution inflammatory syndrome in HIV-infected patients. HIV/AIDS (Auckland, N Z). 2015;7:49–64.
- Bhatt VR, Shostrom V, Gundabolu K, et al. Utilization of initial chemotherapy for newly diagnosed acute myeloid leukemia in the USA. Blood Adv. 2018;2(11):1277–1282.
- Hansen BA, Ø W, Ø B, et al. Febrile Neutropenia in Acute Leukemia. Epidemiology, Etiology, Pathophysiology and Treatment. Mediterr J Hematol Infect Dis. 2020;12(1):e2020009.
- Terwilliger T, Abdul-Hay M. Acute lymphoblastic leukemia: a comprehensive review and 2017 update. Blood Cancer J. 2017;7(6):e577.
- Pagano L, Caira M, Candoni A, et al. The epidemiology of fungal infections in patients with hematologic malignancies: the SEIFEM-2004 study. Haematol. 2006;91(8):1068–1075.
- Montassier E, Gastinne T, Vangay P, et al. Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment Pharm Ther. 2015;42(5):515–528.
- Robinson S, Peterson CB, Sahasrabhojane P, et al. Observational Cohort Study of Oral Mycobiome and Interkingdom Interactions over the Course of Induction Therapy for Leukemia. mSphere. 2020;5(2):e00048–20.
- Roesner LM, Werfel T, Heratizadeh A. The adaptive immune system in atopic dermatitis and implications on therapy. Expert Rev Clin Immunol. 2016;12(7):787–796.
- Boguniewicz M, Leung DY. Atopic dermatitis: a disease of altered skin barrier and immune dysregulation. Immunol Rev. 2011;242(1):233–246.
- Faergemann J. Atopic dermatitis and fungi. Clin Microbiol Rev. 2002;15:545–563.
- Kortekangas-Savolainen O, Lammintausta K, Kalimo K. Skin prick test reactions to brewer’s yeast (Saccharomyces cerevisiae) in adult atopic dermatitis patients. Allergy. 1993;48(3):147–150.
- Oliveira A, Sodré CS, Ferreira DC, et al. Oral aspects identified in atopic dermatitis patients: a literature review. Open Dent J. 2018;12:424–434.
- Wheeler ML, Limon JJ, Underhill DM. Immunity to commensal fungi: detente and disease. Annu Rev Pathol. 2017;12:359–385.
- Javad G, Taheri SM, Hedayati MT, et al. Evaluation of candida colonization and specific humoral responses against candida albicans in patients with atopic dermatitis. Biomed Res Int. 2015. DOI:10.1155/2015/849206.
- Wickes BL, Wiederhold NP. Molecular diagnostics in medical mycology. Nat Commun. 2018;9:5135.
- Kozel TR, Wickes B. Fungal diagnostics. Cold Spring Harb Perspect Med. 2014;4:a019299.
- Temfack E, Kouanfack C, Mossiang L, et al. Cryptococcal antigen screening in asymptomatic HIV-infected antiretroviral native patients in Cameroon and evaluation of the new semi-quantitative BioSynex Crypto PS test. Front Microbiol. 2018;9:409.
- Frau A, Kenny JG, Lenzi L, et al. DNA extraction and amplicon production strategies deeply inffluence the outcome of gut mycobiome studies. Sci Rep. 2019;9:9328.
- Galloway-Peña JR, Kontoyiannis DP. The gut mycobiome: the overlooked constituent of clinical outcomes and treatment complications in patients with cancer and other immunosuppressive conditions. PLoS Pathog. 2020;16(4):e1008353.
- Moher D, Liberati A, Tetzlaff J, et al. The PRISMA group. preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6. DOI:10.1371/journal.pmed.1000097.
- Arendorf TM, Walker DM. The prevalence and intra-oral distribution of Candida albicans in man. Arch Oral Biol. 1980;25:1–10.
- Mukherjee PK, Chandra J, Retuerto M, et al. Oral mycobiome analysis of HIV-infected patients: identification of pichia as an antagonist of opportunistic fungi. PloS Pathog. 2014;10. DOI:10.1371/journal.ppat.1003996.
- Fukui Y, Kotaro A, Yoshikazu I, et al. The palatine tonsil bacteriome, but not the mycobiome, is altered in HIV infection. BMC Microbiol. 2018;18:127.
- Vijendran LCP, Verma R, Hazra N, et al. A comparative study of the various patterns of oro-cutaneous fungi and their sensitivity to anti fungals between HIV patients and normal healthy individuals. Med J Armed Forces India. 2018;75:50–57.
- Li YY, Chen WY, Li X, et al. Asymptomatic oral yeast carriage and antifungal susceptibility profile of HIV-infected patients in Kunming, Yunnan Province of China. BMC Infect Dis. 2013;13:46.
- Hager CL, Ghannoum MA. The mycobiome in HIV. Curr Opin HIV AIDS. 2018;13(1):69–72.
- Annavajhala MK, Khan SD, Sullivan SB, et al. Oral and gut microbial diversity and immune regulation in patients with HIV on antiretroviral therapy. mSphere 2020;5(1):e0079819.
- Das Chagas MS, Portela MB, Cerqueira DF, et al. Reduction of Candida species colonization in the oral cavity of children infected with human immunodeficiency virus after dental treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;108. DOI:10.1016/j.tripleo.2009.04.038.
- Junqueira JC, Vilela SFG, Rossoni RD, et al. Oral colonization by yeasts in HIV-positive patients in Brazil. Rev Inst Med Trop. 2012;54:17–24.
- Moris DV, Melhem MSC, Martins MA, et al. Prevalence and antifungal susceptibility of Candida parapsilosis complex isolates collected from oral cavities of HIV-infected individuals. J Med Microbiol. 2012;61:1758–1765.
- Mukherjee PK, Chandra J, Retuerto M, et al. Dysbiosis in the oral bacterial and fungal microbiome of HIV- infected subjects is associated with clinical and immunologic variables of HIV infection. Plos One. 2018;13(7):e0200285.
- Dos Santos Abrantes PM, McArthur CP, Africa CWJ. Multi-drug resistant (MDR) oral Candida species isolated from HIV-positive patients in South Africa and Cameroon. Diagn Microbiol Infect Dis. 2013;79:222–227.
- Merenstein D, Haihong H, Wang C, et al. Colonization by Candida Species of the Oral and Vaginal Mucosa in HIV-Infected and Noninfected Women. AIDS Res Hum Retrov. 2013;29(1):30–34.
- Sharifzadeh A, Khosravi AR, Shokri H. Oral microflora and their relation to risk factors in HIV + patients with oropharyngeal candidiasis. J Mycol Med. 2013;23:105–112.
- Jiang L, Yong X, Li R, et al. Dynamic analysis of oral Candida carriage, distribution, and antifungal susceptibility in HIV-infected patients during the first year of highly active antiretroviral therapy in Guangxi, China. J Oral Pathol Med. 2014;43:696–703.
- Thanyasrisung P, Kesakomol P, Pipattanagonvit P, et al. Oral Candida carriage and immune status in Thai human immunodeficiency virus-infected individuals. J Med Microbiol. 2014;63:753–759.
- Menezes RP, Borges AS, Araujo LB, et al. Related factors for colonization by Candida species in the oral cavity of HIV-infected individuals. Ver Inst Med Trop. 2015;57:413–419.
- Lourenço AG, Ribeiro AERA, Nakao C, et al. Oral Candida spp carriage and periodontal diseases in HIV-infected patients in Ribeirão Preto, Brazil. Rev Inst Med Trop. 2017;59:e29.
- De Mendonça RM, Araújo MD, Levy CE, et al. Oral mucositis in pediatric acute lymphoblastic leukemia patients: evaluation of microbiological and hematological factors. Pediatr Hematol Oncol. 2015;32(5):322–330.
- De Mendonça RM, De Araújo M, Levy CE, et al. Prospective evaluation of HSV, Candida spp., and oral bacteria on the severity of oral mucositis in pediatric acute lymphoblastic leukemia. Support Care Cancer. 2012;20:1101–1107.
- Akdis CA, Akdis M, Bieber T, et al. European Academy of Allergology and Clinical Immunology/American Academy of Allergy Asthma and Immunology: diagnosis and treatment of atopic dermatitis in children and adults: european Academy of Allergology and Clinical Immunology/American Academy of Allergy, Asthma and Immunology/PRACTALL Consensus Report. J Allergy Clin Immunol. 2006;118:152–169.
- Fonacier LS, Dreskin SC, Leung DYM. Allergic skin diseases. J Allergy Clin Immunol. 2010;125:138–149.
- Incorvaia C, Frati F, Verna N, et al. Allergy and the skin. Clin Exp Immunol. 2008;153:27–29.
- Luis A, Schaub B. Lesson from the farm environment. Curr Opin Allergy Clin Immunol. 2012;12:153–163.
- Glatz M, Bosshard P, Grendelmeier PS. The role of fungi in atopic dermatitis. Immunol Allergy Clin. 2017;37:63–74.
- Feja KN, Wu F, Roberts K, et al. Risk factors for candidemia in critically ill infants: a matched case-control study. J Pediatr. 2005;147(2):156–161.
- Williams DW, Lewis MA. Isolation and identification of candida from the oral cavity. Oral Dis. 2000;6:3–11.
- Mane A, Kulkami A, Risbud A. Biofilm production in oral Candida isolates from HIV-positive individuals from Pune, India. Mycoses. 2012;56(2):182–186.
- Silva JO, Candido RC. Avaliação do sistema API20C AUX na identificação de leveduras de interesse clínico. Rev Soc Bras Med Trop. 2005;38(3):261–263.
- Blignaut E, van Heerden WF. Molecular and histological association between candida albicans from oral soft tissue and carious dentine of HIV-positive children. Mycopathologia. 2015;180:193–201.
- Mushi MF, Mitsemitika CI, Bader O, et al. High Oral Carriage of Non-albicans Candida spp. among HIV-infected individuals. Int J Infect Dis. 2016;49:185–188.
- Forth FL, Hoper D. Highly efficient library preparation for Ion Torrent sequencing using Y-adapters. Biotechniques. 2019;67(5):229–237.
- Carvalho MCCG, Silva DCG. Sequenciamento de DNA de nova geração e suas aplicações na genômica de plantas. Ciênc Rural. 2010;40(3):735–744.
- Amjad M. An overview of the molecular methods in the diagnosis of gastrointestinal infectious diseases. Int J Microbiol. 2020. DOI:10.1155/2020/8135724
- Diaz PI, Hong BY, Dupuy AK, et al. Mining the oral mycobiome: methods, components, and meaning. Virulence. 2017;8(3):313–323. . 1252015
- Huseyin CE, O’Toole PW, Cotter PD, et al. Forgotten fungi-the gut mycobiome in human health and disease. FEMS Microbiol Rev. 2017;41(4):479–511.
- Rhoads DD, Wolcott RD, Sun Y, et al. Comparison of culture and molecular identification of bacteria in chronic wounds. Int J Mol Sci. 2012;13(3):2535–2550.
- Low CY, Rotstein C. Emerging fungal infections in immunocompromised patients. F1000 Med Rep. 2011;3:14.
- Rees JA. Regulation and private participation in the water and sanitation sector. Nat Resour Forum. 1998;22:95–105.
- Denning DW, Kibbler CC, Barnes RA. British society for medical mycology proposed standards of care for patients with invasive fungal infections. Lancet Infect Dis. 2003;3:230–240.
- Garey KW, Rege M, Manjunath PP, et al. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: a multi-institutional study. Clin Infect Dis. 2006;43:25–31.
- Nolla-Salas J, Sitges-Serra A, León-Gil C, et al. Candidemia in non-neutropenic critically ill patients: analysis of prognostic factors and assessment of systemic antifungal therapy. Intensive Care Med. 1997;23:23–30.
- Qian J, Cutler JE, Cole RB, et al. MALDI-TOF mass signatures for differentiation of yeast species, strain grouping and monitoring of morphogenesis markers. Anal Bioanal Chem. 2008;392:439–449.
- Dhiman N, Hall L, Wohlfiel SL, et al. Performance and cost analysis of matrix-assisted laser desorption ionization–time of flight mass spectrometry for routine identification of yeast. J Clin Microbiol. 2011;49:1614–1616.
- Alanio A, Beretti JL, Dauphin B, et al. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for fast and accurate identification of clinically relevant Aspergillus species. Clin Microbiol Infect. 2011;17:750–755.
- De Carolis E, Osteraro B, Lass-Flörl C, et al. Species identification of Aspergillus, Fusarium and Mucorales with direct surface analysis by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect. 2011;18(5):475–484.
- Bader O, Weig M, Thaverne-Gadhwal L, et al. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect. 2011;17:1359–1365.
- Wang Z, Nilsson RH, James TY. Future perspectives and challenges of Fungal Systematics in the Age of Big Data. In Li, De-Wei editors. Biology of Microfungi. CT: Springer International Publishing; 2006. 25–46. DOI:10.1007/978-3-319-29137-6
- Lutzoni F, Kauff F, Cox CJ, et al. Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. Am J Bot. 2004;91:1446–1480.
- Miotto NML, Yurgel LS, Cherubini K, et al. Métodos laboratoriais de identificação do fungo Candida sp. Revista Da Faculdade De Odontologia De Passo Fundo. 2004;9(1):27–33.
- Zhou KM, Lokate M, Deurenberg RH, et al. Use of whole-genome sequencing to trace, control and characterize theregional expansion of extended-spectrum β-lactamase producing ST15 Klebsiella pneumoniae. Sci Rep. 2016;6:20840.
- Edwards DJ, Holt KE. Beginner’s guide to comparative bacterial genome analysis using next-generation sequence data. Microb Inform Exp. 2013;3(1):2.
- Gillevet PM. Multitag Sequencing and Ecogenomic Analysis. U.S. Patent No 8,603,749. Washington, DC: US Patent and Trademark Office; 2006.
- Metzker M. Sequencing technologies — the next generation. Nat Rev Genet. 2010;11:31–46.
- Alanazi H, Semiali A, Chmielewski W, et al. E-Cigarettes Increase Candida albicans Growth and Modulate its Interaction with Gingival Epithelial Cells. Int J Environ Res Public Health. 2019;16:294.
- Mokeem S, Abduliabbar T, Al-Kheraif AA, et al. Oral Candida carriage among cigarette-and waterpipe-smokers, and electronic cigarette users. Oral Dis. 2019;25:319–326.
- Muzurovic S, Hukic M, Babajic E, et al. The relationship between cigarette smoking and oral colonization with Candida species in healthy adult subjects. Med Glas. 2013;10(2):397–399.
- Darwazeh AMG, Dwari ZNA, Al-Zwari AAW. The relationship between tobacco smoking and oral colonization with candida species. J Contemp Dent Pract. 2010;11(3):17–24.
- Becker T, Porat D, Gorsky M. The association between smoking habits and candida in the oral cavity. Int J Dent Oral Health. 2015;1:2.
- Eigner U, Holfelder M, Oberdorfer K, et al. Performance of a matrix-assisted laser desorption ionization time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin Lab. 2009;55(7/8):289–296.
- Angeletti S. Matrix assisted laser desorption time of flight mass spectrometry (MALDI-TOF MS) in clinical microbiology. J Microbiol. 2017;138:20–29.
- Barros NEF, Oliveira EMM, Marin VA. Aplicabilidade da metodologia de reação de polimerase em cadeia em tempo real na determinação do percentual de organismos geneticamente modificados em alimentos. Rev Nutr. 2008;21(1):85–92.
- Matsumura N, Aiba S, Tanaka M, et al. Comparison of immune reactivity profiles against various environmental allergens between adult patients with atopic dermatitis and patients with allergic respiratory diseases. Acta Derm Venereol. 1997;77(5):388–391.
- Diez B, Pedrós-Alió C, Marsh TL, et al. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. App Environ Microbiol. 2001;67:2942–2951.
- Krsek M, Wellington EM. Comparison of different methods for the isolation and purification of total community DNA from soil. J Microbiol Methods. 1999;39(1):1–16.
- Ovreas L. Population and community level approaches for analyzing microbial diversity in natural environments. Ecol Lett. 2000;3:236–251.
- Tsiodras S, Kelesidis T, Kelesidis I, et al. Human infections associated with wild birds. J Infect. 2008;56(2):83–98.