ABSTRACT
Bacteria usually live in complex environments, sharing niche and resources with other bacterial species, unicellular eukaryotic cells or complex organisms. Thus, they have evolved mechanisms to communicate, to compete and to adapt to changing environment as diverse as human tissues, animals or plants. Understanding the molecular mechanisms underlying these adaptation processes is therefore of primary importance for epidemiology and human health protection, and was the focus of a Current Trends in Biomedicine workshop organized by the International University of Andalucia in late October 2015 in Baeza (Spain). The topic was covered by complementary sessions: (i) interbacterial communication and competition that enable a better access to nutrients or a more efficient colonization of the ecological niche, (ii) adaptation of intracellular pathogens to their host, focusing on metabolic pathways, adaptive mechanisms and populational heterogeneity, and (iii) adaptation of animal and plant pathogens as well as plant-associated bacteria to a plant niche. This workshop emphasized the broad repertoire of mechanisms and factors bacteria have evolved to become efficient pathogens.
Introduction
Bacterial adaptation to its environment, considering the environment in its broad sense as every niche in which bacteria have to survive, is a complex issue of undisputed relevance for fields such as ecology, biotechnology, crop protection and biomedicine. While this topic has been underestimated for decades, it is now evident that bacteria have evolved a vast diversity of mechanisms to adapt quickly to new environments and new conditions. These mechanisms are critical for colonization, access to nutrients, and in the case of bacterial pathogens, for the efficiency of the infection process. Interestingly, recent reports have highlighted how bacteria use common molecular mechanisms to achieve adaptation in different environments and toward different ends.Citation1-4 Usually, environments are very complex and bacteria have to cope within multi-species microbiota and consortia. Therefore these studies also emphasized how the analysis of adaptive mechanisms to any given environment became fastidious when the microbiota has also to be taken into account. This additional complexity springs from 2 fundamental aspects: communication and competition between neighboring bacteria, and sensing, signaling and providing the adequate regulatory response to the presence of other bacteria or host cells.Citation4-8 The study of the evolution of the molecular traits involved in all these bacterial processes has evidenced the impact of horizontal gene transfer (HGT) as a major event for the acquisition of new functions in bacteria.Citation9 It also revealed how stress conditions such as those encountered when facing host defenses, antibiotic pressure and other environmental cues promote events leading to decision making, HGT or to bacterial warfare. Thus, understanding different seemingly specific molecular mechanisms involved in interacting with a given environment is relevant to understand any adaptation process. Interestingly, studying bacterial adaptation to fluctuating environments also revealed the phenotypic heterogeneity of the population, and how the diversity between individuals of the same species allow a rapid adaptation and how adapted clones emerge and maintain.Citation3,10 The diversity and the complexity of these issues led us to gather different scientific communities to address a central and identical biological question: What drives the adaptation of bacterial pathogens? This key question constituted the scaffold of a “Current Trends in Biomedicine” workshop recently organized by the International University of Andalucia (UNIA) in Baeza (Spain) and entitled “Adaptation and communication of bacterial pathogens.” This workshop garnered 45 participants from the United States and various countries in Europe (Austria, Denmark, France, Germany, Italy, Spain, United Kingdom) (), including 14 invited speakers. Twenty-six participants presented posters and 10 of them were selected for short-talks. The workshop was divided in 5 sessions describing communication and adaptation between bacterial species, between intracellular bacteria and their hosts, and between bacteria and plant cells. The discussions emphasized the commonalities and the diversity of mechanisms deployed by bacterial pathogens to reach their final goal: the success of the infection process.
Session 1: Inter-bacterial communication, exchange and competition
The first session dealt with bacterium-bacterium interactions as bacteria do not live alone but rather in complex communities. When thriving in their environment, bacteria have to cope with many other species and must therefore collaborate or compete to access nutrients or to colonize more efficiently the ecological niche. There is also a need to discriminate between siblings and real competitors. The talks covering this session contributed to a better understanding of the molecular mechanisms leading to the adaptive dynamics of a bacterial population and the mutualistic and competitive behaviors between bacterial species.
Interspecies interactions could be synergistic. Several mechanisms have evolved for bacterial communication and collaboration, relying on complex regulatory networks, the production of aggregative substances and the transfer or exchange of material between cells (). These mechanisms, such as quorum sensing, biofilm formation or metabolic cooperation, lead to the development of beneficial traits for the community and explain long-term persistence of bacterial strains in the environment or in complex niches such as the human body.Citation11 The session began with the presentation of Søren Molin (Technical University of Copenhagen, Danemark). Molin and co-workers investigated the evolutionarily diversity and adaptative dynamics of Pseudomonas aeruginosa by sequencing 700 genomes from hospital isolates covering a 40-year period. He described how P. aeruginosa evolved to adapt to and invade the human airways. They found 52 patho-adaptation mutations affecting transcriptional regulators, antibiotic resistance traits and cell wall and lipopolysaccharides components, allowing the conversion from naïve to adapted strains. The most adapted clone could then be transmitted from patient-to-patient.Citation12 Maite Echeverz (Agrobiotechnology Institute, Public University of Navarra, Spain), a young researcher from the group of Iñigo Lasa presented a short-talk on the special traits that cellulose and the β-1,6-linked N-acetylglucosamine exopolysaccharides confer to bacterial biofilm. She further showed how these exopolysaccharides influence biofilm resistance to various stresses and virulence. Finally, Carolina Palancia-Gándara (University of Cantabria, Santander, Spain), a young researcher from the group of Fernando de la Cruz, introduced how bacteria exchange genetic material via conjugation. She convinced us that plasmid transfer is the main mechanism for dissemination of antibiotic resistance genes and that there is a need for developing conjugation inhibitors. She provided evidence that natural and synthetic unsaturated fatty acids such as linoleic, 2-hexadecynoic and tanzawaic acids are potent inhibitors of various plasmid transfer systems.Citation13,14
Figure 2. Selected molecular determinants of inter-bacterial interactions. Beneficial interactions include the exchange of material such as outer membrane components, metabolites or intracellular content and the transfer of plasmid DNA to recipient cells by conjugation. Biofilm formation and cell aggregation involve many determinants such as exopolysaccharides and Type IV pili, and allow an increased protection against antibiotics or physical stresses. Bacterial competition involves the release of antagonistic molecules, peptides and proteins or the direct delivery of toxin effectors by dedicated machineries, such as Type VI secretion and contact-dependent growth inhibition systems.
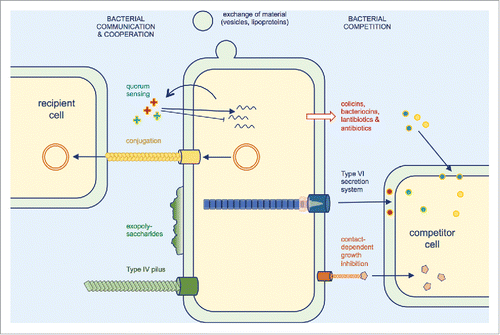
The second part of this session was dedicated to bacterial competition. Bacteria often live in complex multi-species communities and have to compete for the limited resources. They are therefore subjected to antagonism behaviors, as recently evidenced by following how bacteria grow in mixed cultures.Citation15,16 Although competition between bacterial species has been underestimated for decades it recently garnered attention with the discovery of dedicated mechanisms or machineries that create direct cell damages to the competitor or poison the competitor by the delivery of anti-bacterial toxins (). However, bacterial competition does not only affect bacterial fate and multispecies communities, but also indirectly influences the pathogenesis outcome. The various mechanisms have been described in the 4 last talks of the session. David Low (University of Santa Barbara, CA, USA) gave a very dynamic presentation on the contact-dependent growth inhibition (CDI) mechanism. This relies on the delivery of anti-bacterial toxins, called CdiA, by a sub-family of Type V secretion, 2-partner secretion.Citation17 Although the mechanism on how these filamentous toxins are transported to the cell exterior of the attacker cell is well conserved, his talk emphasized the broad variability of strategies these toxins use to recognize target cells and to parasitize target cell components to reach their final destination. Particularly, he presented data that defined the molecular and structural determinants of CdiA binding to its receptor BamA, as well as a mutagenesis study to identify target cell components required for efficient CdiA translocation.Citation18,19 The 3 last talks of the session were dedicated to the Type VI secretion system. This secretion apparatus is composed a bacteriophage-derived contractile tail used to propel an arrow-like structure that punctures the target cell and delivers toxin effectors.Citation20,21 Eric Cascales (CNRS/Aix-Marseille Université, France) showed how this contractile apparatus is anchored to the cell envelope. Using a combination of genetic, biochemical, structural and fluorescence microscopy approaches, they defined the components and the architecture of this membrane complex. They showed that this complex is the first to be assembled and serves both as a docking station for the tail and as a channel for the passage of the arrow. He finally described the negative-stain electron microscopy structure of the 1.7-MDa membrane complex at 12-Å resolution obtained in collaboration with Rémi Fronzes's group.Citation22 Laura Nolan (Imperial College, London, UK), a post-doctoral researcher from the group of Alain Filloux, presented recent data using transposon-directed insertion sequencing to identify toxins delivered by the H1-T6SS from P. aeruginosa. Her approach was validated by the identification of known effectors and uncovered potential new toxins. S. Brook Peterson (University of Washington, Seattle, WA, USA), a research scientist from the group of Joseph Mougous, demonstrated how Type VI-mediated toxin delivery influences the composition of microbial communities. In collaboration with Andrew Goodman's group, she specifically showed that Bacteroidetes are equipped with a surprisingly high diversity of anti-bacterial toxins that are involved in maintaining the symbiotic relationship with the mammalian gut.Citation23
Sessions 2 and 3: Adaptation of intracellular bacteria to their hosts
The second and third sessions of the workshop were dedicated to the description and the characterization of adaptive mechanisms of intracellular pathogens within their hosts. Deciphering such mechanisms used by these bacteria to cope with their stressful environments is a complex issue that requires a better understanding of the molecular basis of bacterial pathogenicity. Over the last 2 decades, massive efforts have been undertaken to reach a better knowledge of toxin production, quorum sensing and functions of secretion systems. More recently, a renewed interest in metabolism and adaptation inside the host has emerged, with an intensification of studies dealing with the characterization of non-dividing bacteria (). These topics were tackled during these 2 sessions, highlighting several regulatory mechanisms and metabolic pathways used by bacteria during their intracellular life to succeed in their infectious process.
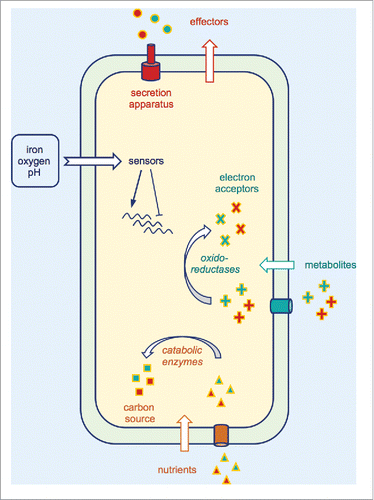
Figure 3. Selected adaptive mechanisms used by bacterial pathogens to survive within their hosts. These mechanisms include secretion and delivery of effectors which interfere with the host cytoskeleton and immune signaling. pH-, iron- and oxygen-dependent bacterial sensors can be activated to modulate expression of their regulons, leading to gene expression reprogramming and favoring bacterial adaptation. Dedicated bacterial enzymes can be used (i) to metabolize nutrients and (ii) to reduce or oxidize metabolites present in the host environment, both reactions conferring to the pathogen, an advantage over the competing microbiota.
Session 2: Regulatory mechanisms of adaptation of intracellular bacteria
David Holden (Imperial College, London, UK) chaired the second session and gave the first presentation. He described the characterization of 2 effectors translocated by the Salmonella Pathogenicity Island 2 Type III secretion system (SPI-2 T3SS), SseF and SseG. Both proteins are located in the Salmonella-containing vacuole (SCV) membrane and interact with each other.Citation24 SseF and SseG were shown to mediate association of SCVs with the Golgi network within epithelial cells. A yeast 2-hybrid screen identified a host cell Golgi protein that interacts with both SseG and SseF. Depletion of the host protein prevented vacuoles-containing wild-type bacteria from interacting stably with the Golgi network. Finally, Holden and coworkers showed that SseFG complex formation could be abolished by random mutagenesis of SseG, and proposed a model that partly explains how SseF and SseG work together. The second presentation by Renée Tsolis (University of California Davis, USA) tackled the question on how the innate immune system senses a Type IV secretion system (T4SS) effector of Brucella abortus called VceC. B. abortus replicates in infected macrophages within an endoplasmic reticulum (ER)-associated compartment. Formation of this compartment require an active T4SS. T4SS-dependent transfer of VceC into macrophages perturbs the function of the endoplasmic reticulum and activates the host cell's unfolded protein response via the ER sensor IRE1, hence leading to the secretion of IL-6. This work suggested that the IRE1 pathway of ER stress sensing serves an innate immune function into the host cell that senses B. abortus infection.Citation25,26 Next, Josep Casadesús (Sevilla University, Spain) presented 2 examples of non-mutational preadaptations. He began by describing a random preadaptation mechanism - Salmonella exposure to bile - that triggers the RpoS-dependent general stress response and increases bile resistance. RpoS expression permits survival of certain cells in the presence of bile, and a positive feedback loop sustains or even amplifies the RpoS response, giving rise to a bile-resistant population.Citation27 Casadesús' second example dealt with programmed preadaptation. The S. enterica opvAB operon encodes 2 cytoplasmic membrane proteins that alter lipopolysaccharide O-antigen chain length and confers resistance to bacteriophages that use the O-antigen as receptor. Because expression of opvAB undergoes phase variation, S. enterica populations contain a mixture of opvABON and opvABOFF cells. Infection of Salmonella with a virulent phage kills the opvABOFF subpopulation and selects the opvABON subpopulation, preadapting these bacteria to survive phage challenge in a reversible manner.Citation28 Next, Olivier Espéli (Collège de France, Paris, France) gave a talk dedicated to the adaptation of the adherent-invasive E. coli LF82 strain inside mature phagolysosomes. This strain has been isolated from a Crohn's disease patient and has the ability to invade epithelial cells and to proliferate within macrophages. Interestingly, LF82 does not detoxify its environment and therefore induces many bacterial stress responses. As a consequence of this challenging environment, a small subset of bacteria retains the capacity to replicate while others either form non replicating persisters or are killed. This session was closed by Francisco García-del Portillo (CSIC Madrid, Spain) who detailed a suicide strategy involving accumulation of endomembranes to control Salmonella proliferation inside the fibroblast, a cell type in which the pathogen establishes a long lasting persistent infection.Citation29 His presentation illustrated how an intracellular pathogen can communicate to the host triggering defenses from inside the infected cell to mount “a suicide program.”
Session 3: Metabolism of intracellular bacteria
The third session started with a talk from David Russell (Cornell University, USA) who aimed to understand how the host environment shapes the physiology of Mycobacterium tuberculosis (Mtb). By conducting an extensive, unbiased chemical screen to identify small molecules that inhibit Mtb metabolism within macrophages, Russell and co-workers identified a significant number of novel compounds that limit Mtb growth in macrophages and in medium containing cholesterol as the major carbon source. Based on this observation, they developed a chemical-rescue strategy to identify compounds that target metabolic enzymes involved in cholesterol metabolism. These chemical probes represent new classes of inhibitors that target metabolic pathways required to support growth of Mtb in its host cell.Citation30 In addition, studies into the mode of action of existing frontline drugs on intracellular Mtb revealed how host-derived stresses contribute to the rise of a drug tolerant phenotype. This does however represent an opportunity for the identification of synergistic inhibitors that could ameliorate existing drug therapies. Next, Laurent Aussel (Aix-Marseille Université, France) showed how Salmonella enterica modulates its pathogenesis in response to iron and oxygen availability in the environment. The ISC machinery, involved in iron-sulfur [Fe-S] protein biogenesis, was demonstrated to play a central role in Salmonella virulence through the ability of IscR - a transcriptional regulator carrying a [2Fe-2S] cluster - to downregulate SPI-1 T3SS gene expression. Iron starvation and oxidative stress being detrimental for [Fe-S] enzyme biogenesis, a balance occurs between IscR apo-form (clusterless) and its holo-form. This model represents a novel adaptive mechanism used by Salmonella to favor its infectivity in the gut, where oxygen is rare and iron abundant, whereas SPI-1 T3SS would be repressed in macrophages to reduce energetic expenses. The following presentation was given by Andreas Bäumler (University of California Davis, USA) and began by an overview of the general metabolic pathways used by Salmonella in the gut. Previous works from his lab showed that inflammation-derived nutrients available in this new niche support a bloom of Salmonella serovars in the gut lumen, ensuring transmission of the pathogen to the next susceptible host by the fecal-oral route.Citation31,32 In his talk, Bäumler showed that a set of 469 genes involved in the central anaerobic metabolism was degrading in genomes of Salmonella serovars exclusively associated with extraintestinal infections but remained intact in genomes of Salmonella serovars associated with human gastroenteritis.Citation31 This metabolic network identified by comparative genome analysis provides clues about the strategies for nutrient acquisition and utilization that are characteristic of gastrointestinal pathogens and conferring to Salmonella a “winning metabolic strategy” to edge out competing microbes in the inflamed intestine (). The session was closed by Julie Viala (CNRS/Aix-Marseille Université, France) who addressed the role of the acyl carrier protein-like IacP protein in Salmonella enterica. Its corresponding gene as well as those encoding the T3SS are all localized within the SPI-1 gene cluster.Citation33 Her work showed that the SPI-1 T3SS is post-translationally modified according to a process that involves IacP, leading to the optimization of the pore-forming activity of the machinery.
Sessions 4 and 5: Bacterial adaptation to the plant environment
The fourth and fifth sessions jointly focused on commonalities and differences between the bacterial adaptation mechanisms involved in distinct lifestyles within the host: pathogens, commensals and symbionts. Recent studies have shown how common bacterial mechanisms can differentially target host processes, rendering the interaction from mutually beneficial to pathogenic. Such differences, so relevant from the point of view of fighting pathogen infections, may prove highly important for the design of antimicrobial compounds and treatments. Currently, such complete comparison between different bacteria-host interactions can only be comprehensively addressed in a plant model, both for the comparatively simpler physiology of plants as hosts, and their potential for genetic analysis and modification. Thus, plants have become model systems for such studies, giving rise to a high number of molecular tools developed for the genetic characterization of the host processes involved in host adaptation, and for the plasticity they display for microbe interactions. Session four focused on the adaptation of plant and animal bacterial pathogens to the plant environment. This session was organized to cover the commonalities and differences found between the mechanisms involved in adaptation of both plant and animal bacterial pathogens to the plant host, and those animal pathogens use to differentially adapt to animal and plant hosts (). This provided us with an interesting and rather unique opportunity to integrate a discussion between plant and animal microbiologists. The fifth and last session of the meeting focused around the adaptation to the plant environment of non-pathogenic bacteria. Insights on the mechanisms involved in the adaptation and interaction with the host or other microorganisms of these beneficial bacteria were presented in this session and were discussed in the context of the previous sessions in which the mechanisms used by their pathogenic counterparts were presented.
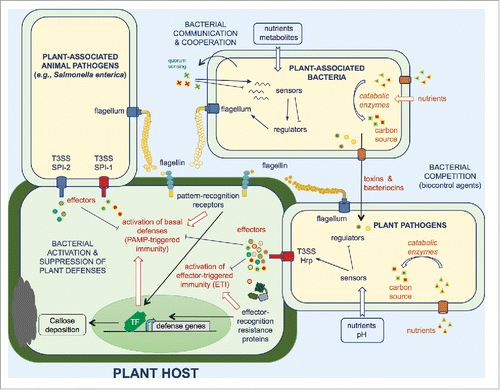
Figure 4. Selected adaptive mechanisms used by animal and plant pathogens as well as plant-associated bacteria to survive within their hosts. These mechanisms include secretion and delivery of effectors which interfere with host immune signaling. Bacterial sensors can be activated in response to different environmental cues such as pH or nutrients, to modulate expression of their regulons, leading to gene expression reprogramming and favoring bacterial adaptation. Dedicated bacterial enzymes can be used (i) to metabolize nutrients and (ii) to modify the host microenvironment, both reactions conferring to the pathogen an advantage over the competing microbiota. Biofilm formation and cell aggregation may also play a role in the adaptation to the plant environment and allow an increased protection against stresses. Bacterial competition involves the release of antagonistic molecules, peptides and proteins or the direct delivery of toxins.
Session 4: Adaptation to the plant environment of phytopathogenic bacteria and animal pathogens
Opening session 4, Adam Schikora (Institute for Epidemiology and Pathogen Diagnostics, JKI Braunschweig, Germany) provided us with the opportunity to compare the molecular determinants required for Salmonella to adapt to a plant host with those required for its adaptation to the animal host.Citation34 Both types of hosts detect Salmonella upon contact, by perceiving flagellin. However, those different hosts perceive distinct domains of flagellin that leads to the activation of basal defenses in both cases. Salmonella requires 2 T3SSs that are necessary for colonisation of and proliferation within plant and animal hosts, however, the role of the individual effector proteins translocated by either system varies depending on the host system. Several effectors translocated by the T3SS encoded by the SPI-2 locus and required for intracellular replication within macrophages, were shown to suppress the plant responses triggered upon recognition of flagellin, whereas one effector translocated by both T3SS was shown to act on MAPK-mediated signaling cascades to suppress plant immunity ().Citation35 Stéphane Genin (Laboratoire des Interactions Plantes-Microorganismes, Castanet-Tolosan, France) presented an evolution experiment in which the phenotypic and genotypic changes happening to the broad-host range quarantine plant pathogenic bacterium Ralstonia solanacearum during its adaptation to tolerant, resistant or susceptible plant hosts, was followed for 350 generations. New isolates were then tested in challenge experiments against the ancestor in different hosts. The results showed that evolution within the hostile environment of a resistant host triggers the acquisition of mutations leading to more competitive, better adapted isolates.Citation36 Sequencing and analysis of the genetic changes associated with the adaptation process provided new information about the genetic determinants involved, and allowed the identification of conserved mutations appearing independently several times in the experiment.Citation36 Carmen Beuzón (IHSM, University of Malaga-CSIC, Spain) presented 2 very different strategies by which the model plant pathogenic bacterium Pseudomonas syringae avoids plant defenses: a stealth strategy, and an active mechanism. In the first case, part of the bacterial population hide themselves from the plant host thanks to a bistable switch that allows them to differentially activate important virulence determinants. These stealthy bacteria are presumably carried along within the plant by their more aggressive counterparts and may adapt better to other environments during different stages of their lifecycle. This strategy is very similar to those employed by animal pathogens such as Salmonella to survive and proliferate within animal hosts, and to those involved in the generation of antibiotic persisters. Active suppression involves a more direct mechanism by which the pathogen uses a T3SS-translocated effector to modify a plant protein involved in activating all levels of defense. Modification by the effector renders the plant target less efficient thus lowering its defenses. Finally, the session was closed by Eloy Caballo, a young researcher from Cayo Ramos' laboratory (IHSM, University of Malaga-CSIC, Spain), who presented a genomic analysis aimed to identify specific genome regions involved in adapting to woody hosts. One of the chromosomal loci they identified is involved in the modification of lignin-related compounds and its mutation causes a decreased virulence toward woody but not herbaceous host models.
Session 5: Adaptation to the plant environment of plant-associated bacteria
Session five set off with a presentation from Marta Martín (Depto. Biología, Universidad Autónoma de Madrid, Spain) on the molecular mechanisms involved in the adaptation of non-pathogenic bacteria to the plant root system. Adaptation experiments led them to identify stable genetic variants with faster motility, capable of outcompeting their ancestor. This genetic diversification is achieved by the activity of site-specific recombinases that results in an increased rate of phase variation of different traits. Their study provides evidence on the relationship between motility and colonization ability, and on the quantitative multigenic nature of bacterial motility. Then, Robert Jackson (School of Biological Science, University of Reading, UK) described the evolutionary changes that occur in the plant-associated bacterium Pseudomonas fluorescens when subjected to nutrient stress and carrying a motility defect. These genetic changes affect the regulatory pathway involved in controlling Nitrogen assimilation in bacteria and led to a change in regulator specificity that results in the re-activation of the flagellar system through an unorthodox signaling cascade.Citation37 Robert's results provided evidence of how regulatory circuits can be exploited during the adaptation process by accumulating genetic changes leading to their rewiring.Citation37 Mateo San José, an early career researcher from his team presented his research on the pathogenic bacterium P. viridiflava, and particularly on the identification of a novel genetic locus with hallmarks of a new quorum sensing system that is essential for virulence in plants. The session closed with a short talk by another young researcher, Tanya Arseneault (School of Biological Sciences, University of Reading, UK), who presented past work (University of Moncton, Canada) on P. fluorescens as a biocontrol agent against the potato pathogen Streptomyces scabies and the link between the production of a toxin with antibiotic properties and biocontrol capacity, within the controlled laboratory conditions against the more complex adaptation process that takes place in field conditions.Citation38
Concluding remarks
The talks, discussions and poster presentations of the workshop emphasized how bacteria adapt to the different conditions they encounter during their life cycle to grow, colonize specific niches and circumvent the attacks of predators or the defenses of hosts to be efficient pathogens. Although bacteria use a broad repertoire of regulatory mechanisms and virulence factors, it is now clear that the efficiency of the infection process does not only involve secretion of toxins but rather is dependent on the adaptation of bacteria to their environment, on the communication between individuals as well as on the outcome of competition behaviors between bacterial species in the same niche. Variations in environmental conditions lead the pathogen to activate multiple sensors, modulating gene expression and allowing bacteria to quickly adjust their metabolism, re-program their defenses and the arsenal of secretion systems and effector proteins. The workshop stressed a number of mechanisms that are critical for the infection process and that have been underestimated, including bacterial competition and the heterogeneity of the bacterial population. A species, in its environment, should not be considered as a community of identical bacteria, but rather as a multitude of individuals with specific properties and behaviors. This heterogeneity, which results from diverse expression programs based on bistability and phase variation within the population evolved from the same progeny, is a pre-adaptive state that allows the selection of the most effective individuals for the infection process. All these notions were broadly discussed during the workshop and emphasized how bacteria, although considered as ‘simple organisms’, have developed smart mechanisms to survive within the environment.
The workshop was held in Baeza in the province of Jaén in Spain, a city surrounded by olive groves, a beautiful environment in which the participants adapted quickly! The workshop organization included a visit of the city, and the participants discovered the impressive Renaissance buildings and history, particularly the Palacio del Jabalquinto, the Cathedral, and the Ancient University of Baeza where taught the great Spanish poet Antonio Machado.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the science advisory committee of the International University of Andalucia for selecting and funding this workshop, the staff of the International University of Andalucia, and particularly Joaquín Torreblanca López, for the organization of the meeting. We also thank Josep Casadesús for encouragements and support, and all the speakers for giving permission to summarize their talks.
Funding
Agence Nationale de la Recherche provided funding for the Laboratoire d'Ingenierie des Systemes Macromoleculaires on the molecular mechanisms regulating bacterial competition in the form of grant number ANR-14-CE14-0006-02.
References
- West SA, Griffin AS, Gardner A. Evolutionary explanations for cooperation. Curr Biol 2007; 17:R661-72; PMID:17714660; http://dx.doi.org/10.1016/j.cub.2007.06.004
- Knief C, Delmotte N, Vorholt JA. Bacterial adaptation to life in association with plants - A proteomic perspective from culture to in situ conditions. Proteomics 2011; 11:3086-105; PMID:21548095; http://dx.doi.org/10.1002/pmic.201000818
- Bentley SD, Parkhill J. Genomic perspectives on the evolution and spread of bacterial pathogens. Proc Biol Sci 2015; 282:20150488; PMID:26702036; http://dx.doi.org/10.1098/rspb.2015.0488
- Steinert M. Pathogen intelligence. Front Cell Infect Microbiol 2014; 4:8; PMID:24551600
- Blango MG, Mulvey MA. Bacterial landlines: contact-dependent signaling in bacterial populations. Curr Opin Microbiol 2009; 12:177-81; PMID:19246237; http://dx.doi.org/10.1016/j.mib.2009.01.011
- Strassmann JE, Gilbert OM, Queller DC. Kin discrimination and cooperation in microbes. Annu Rev Microbiol 2011; 65:349-67; PMID:21682642; http://dx.doi.org/10.1146/annurev.micro.112408.134109
- Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol 2013; 11:285-93; PMID:23456045; http://dx.doi.org/10.1038/nrmicro2977
- Eisenreich W, Heesemann J, Rudel T, Goebel W. Metabolic host responses to infection by intracellular bacterial pathogens. Front Cell Infect Microbiol 2013; 3:24; PMID:23847769
- Juhas M. Horizontal gene transfer in human pathogens. Crit Rev Microbiol 2015; 41:101-8; PMID:23862575; http://dx.doi.org/10.3109/1040841X.2013.804031
- Ackermann M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol 2015; 13:497-508; PMID:26145732; http://dx.doi.org/10.1038/nrmicro3491
- Elias S, Banin E. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol Rev 2012; 36:990-1004; PMID:22229800; http://dx.doi.org/10.1111/j.1574-6976.2012.00325.x
- Marvig RL, Sommer LM, Molin S, Johansen HK. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet 2015; 47:57-64; PMID:25401299; http://dx.doi.org/10.1038/ng.3148
- Fernández-López R, Machón C, Longshaw CM, Martin S, Molin S, Zechner EL, Espinosa M, Lanka E, de la Cruz F. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology 2005; 151:3517-26; PMID:16272375; http://dx.doi.org/10.1099/mic.0.28216-0
- Getino M, Sanabria-Ríos DJ, Fernández-López R, Campos-Gómez J, Sánchez-López JM, Fernández A, Carballeira NM, de la Cruz F. Synthetic fatty acids prevent plasmid-mediated horizontal gene transfer. MBio 2015; 6:e01032-15; PMID:26330514; http://dx.doi.org/10.1128/mBio.01032-15
- Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 2010; 7:25-37; PMID:20114026; http://dx.doi.org/10.1016/j.chom.2009.12.007
- Brunet YR, Espinosa L, Harchouni S, Mignot T, Cascales E. Imaging type VI secretion-mediated bacterial killing. Cell Rep 2013; 3:36-41; PMID:23291094; http://dx.doi.org/10.1016/j.celrep.2012.11.027
- Ruhe ZC, Low DA, Hayes CS. Bacterial contact-dependent growth inhibition. Trends Microbiol 2013; 21:230-7; PMID:23473845; http://dx.doi.org/10.1016/j.tim.2013.02.003
- Willett JL, Gucinski GC, Fatherree JP, Low DA, Hayes CS. Contact-dependent growth inhibition toxins exploit multiple independent cell-entry pathways. Proc Natl Acad Sci U S A 2015; 112:11341-6; PMID:26305955; http://dx.doi.org/10.1073/pnas.1512124112
- Ruhe ZC, Wallace AB, Low DA, Hayes CS. Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. MBio 2013; 4:e00480-13; PMID:23882017; http://dx.doi.org/10.1128/mBio.00480-13
- Zoued A, Brunet YR, Durand E, Aschtgen MS, Logger L, Douzi B, Journet L, Cambillau C, Cascales E. Architecture and assembly of the Type VI secretion system. Biochim Biophys Acta 2014; 1843:1664-73; PMID:24681160; http://dx.doi.org/10.1016/j.bbamcr.2014.03.018
- Cianfanelli FR, Monlezun L, Coulthurst SJ. Aim, Load, Fire: The Type VI secretion system, a bacterial nanoweapon. Trends Microbiol 2016; 24:51-62; PMID:26549582; http://dx.doi.org/10.1016/j.tim.2015.10.005
- Durand E, Nguyen VS, Zoued A, Logger L, Péhau-Arnaudet G, Aschtgen MS, Spinelli S, Desmyter A, Bardiaux B, Dujeancourt A, et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature 2015; 523:555-60; PMID:26200339; http://dx.doi.org/10.1038/nature14667
- Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 2014; 16:227-36; PMID:25070807; http://dx.doi.org/10.1016/j.chom.2014.07.007
- Deiwick J, Salcedo SP, Boucrot E, Gilliland SM, Henry T, Petermann N, Waterman SR, Gorvel JP, Holden DW, Méresse S. The translocated Salmonella effector proteins SseF and SseG interact and are required to establish an intracellular replication niche. Infect Immun 2006; 74:6965-72; PMID:17015457; http://dx.doi.org/10.1128/IAI.00648-06
- de Jong MF, Starr T, Winter MG, den Hartigh AB, Child R, Knodler LA, van Dijl JM, Celli J, Tsolis RM. Sensing of bacterial type IV secretion via the unfolded protein response. MBio 2013; 4:e00418-12; PMID:23422410; http://dx.doi.org/10.1128/mBio.00418-12
- Celli J, Tsolis RM. Bacteria, the endoplasmic reticulum and the unfolded protein response: friends or foes? Nat Rev Microbiol 2015; 13:71-82; PMID:25534809; http://dx.doi.org/10.1038/nrmicro3393
- Hernández SB, Cota I, Ducret A, Aussel L, Casadesús J. Adaptation and preadaptation of Salmonella enterica to bile. PLoS Genet 2012; 8:e1002459; http://dx.doi.org/10.1371/journal.pgen.1002459
- Cota I, Sánchez-Romero MA, Hernández SB, Pucciarelli MG, García-del Portillo F, Casadesús J. Epigenetic control of Salmonella enterica O-antigen chain length: a tradeoff between virulence and bacteriophage resistance. PLoS Genet 2015; 11:e1005667; PMID:26583926; http://dx.doi.org/10.1371/journal.pgen.1005667
- Núñez-Hernández C, Alonso A, Pucciarelli MG, Casadesús J, García-del Portillo F. Dormant intracellular Salmonella enterica serovar Typhimurium discriminates among Salmonella pathogenicity island 2 effectors to persist inside fibroblasts. Infect Immun 2014; 82:221-32; http://dx.doi.org/10.1128/IAI.01304-13
- VanderVen BC, Fahey RJ, Lee W, Liu Y, Abramovitch RB, Memmott C, Crowe AM, Eltis LD, Perola E, Deininger DD, et al. Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium's metabolism is constrained by the intracellular environment. PLoS Pathog 2015; 11:e1004679; PMID:25675247; http://dx.doi.org/10.1371/journal.ppat.1004679
- Nuccio SP, Bäumler AJ. Comparative analysis of Salmonella genomes identifies a metabolic network for escalating growth in the inflamed gut. mBio 2014; 5:e00929-00914; PMID:24643865; http://dx.doi.org/10.1128/mBio.00929-14
- Rivera-Chávez F, Bäumler AJ. The pyromaniac inside you: Salmonella metabolism in the host gut. Ann Rev Microbiol 2015; 69:31-48; http://dx.doi.org/10.1146/annurev-micro-091014-104108
- Viala JP, Puppo R, My L, Bouveret E. Posttranslational maturation of the invasion acyl carrier protein of Salmonella enterica serovar Typhimurium requires an essential phosphopantetheinyl transferase of the fatty acid biosynthesis pathway. J Bacteriol 2013; 195:4399-405; PMID:23893113; http://dx.doi.org/10.1128/JB.00472-13
- Wiedemann A, Virlogeux-Payant I, Chaussé AM, Schikora A, Velge P. Interactions of Salmonella with animals and plants. Front Microbiol 2015; 5:791; PMID:25653644; http://dx.doi.org/10.3389/fmicb.2014.00791
- Neumann C, Fraiture M, Hernàndez-Reyes C, Akum FN, Virlogeux-Payant I, Chen Y, Pateyron S, Colcombet J, Kogel KH, Hirt H, et al. The Salmonella effector protein SpvC, a phosphothreonine lyase is functional in plant cells. Front Microbiol 2014; 5:548; PMID:25368608
- Guidot A, Jiang W, Ferdy JB, Thébaud C, Barberis P, Gouzy J, Genin S. Multihost experimental evolution of the pathogen Ralstonia solanacearum unveils genes involved in adaptation to plants. Mol Biol Evol 2014; 31:2913-28; PMID:25086002; http://dx.doi.org/10.1093/molbev/msu229
- Taylor TB, Mulley G, Dills AH, Alsohim AS, McGuffin LJ, Studholme DJ, Silby MW, Brockhurst MA, Johnson LJ, Jackson RW. Evolutionary resurrection of flagellar motility via rewiring of the nitrogen regulation system. Science 2015; 347:1014-7; PMID:25722415; http://dx.doi.org/10.1126/science.1259145
- Arseneault T, Goyer C, Filion M. Pseudomonas fluorescens LBUM223 increases potato yield and reduces common scab symptoms in the field. Phytopathology 2015; 105:1311-7; PMID:25961336; http://dx.doi.org/10.1094/PHYTO-12-14-0358-R

