ABSTRACT
Non-toxigenic V. parahaemolyticus isolates (tdh−/trh−/T3SS2−) have recently been isolated from patients with gastroenteritis. In this study we report that the larvae of the wax moth (Galleria mellonella) are susceptible to infection by toxigenic or non-toxigenic clinical isolates of V. parahaemolyticus. In comparison larvae inoculated with environmental isolates of V. parahaemolyticus did not succumb to disease. Whole genome sequencing of clinical non-toxigenic isolates revealed the presence of a gene encoding a nudix hydrolase, identified as mutT. A V. parahaemolyticus mutT mutant was unable to kill G. mellonella at 24 h post inoculation, indicating a role of this gene in virulence. Our findings show that G. mellonella is a valuable model for investigating screening of possible virulence genes of V. parahaemolyticus and can provide new insights into mechanisms of virulence of atypical non-toxigenic V. parahaemolyticus. These findings will allow improved genetic tests for the identification of pathogenic V. parahaemolyticus to be developed and will have a significant impact for the scientific community.
Introduction
Vibrio parahaemolyticus is a human pathogen and the leading global cause of seafood-associated gastroenteritis. This disease is characterised by watery or bloody diarrhoea, vomiting, abdominal cramps, headaches, fever and nausea,Citation1 following the consumption of raw or undercooked seafoods.Citation2–4 Several virulence determinants in V. parahaemolyticus have previously been identified including adhesins (e.g. MAM7), thermostable direct haemolysin (TDH) and TDH related haemolysin (TRH) and 2 Type Three Secretion Systems, T3SS1 and T3SS2. The multivalent adhesion molecule, MAM7 is present on the surface of V. parahaemolyticus and has been shown to be critical for the attachment of the bacteria to eukaryotic cells during the early stages of infection.Citation5 The TDH and TRH haemolysins are cytotoxic and enterotoxic pore forming toxins. The T3SS delivers effector proteins such as VopA, VopL, VopT, VopV and VopCCitation6–10 into eukaryotic cells which subsequently modulate host cell processes. T3SS1 is present in both clinical and environmental isolates of V. parahaemolyticus whilst T3SS2 is predominantly present in clinical isolates and is associated with pandemic strains of V. parahaemolyticus and large outbreaks of disease.Citation11 The tdh, trh and the T3SS2 genes are currently considered to be the main virulence determinants for this pathogen.Citation12
Recently, the isolation of clinical strains that lack the tdh, trh and T3SS2 genes has been reported.Citation13–16 For example, in South Thailand between 2001–2010, 9–10% clinical isolates from patients were identified as non-toxigenic.Citation13,14 In 2010, clinical isolates lacking tdh, trh and T3SS2 genes were isolated from patients affected with acute gastroenteritis in Torino, Italy as a result from the consumption of indigenously produced seafood.Citation15 A number of possibilities have been proposed to explain the apparent association of non-toxigenic isolates with disease. Firstly, the ingestion of a mixed population of bacteria might allow the simultaneous growth of toxigenic and non-toxigenic bacteria and a single isolate per patient may lead to misdiagnosis of the aetiological agent.Citation15,17 Secondly, it is possible that the deletion of virulence genes may have occurred during bacterial growth in vitro or in vivo. For example, it has previously been shown that a clinical V. parahaemolyticus isolate lacking tdh resulted from an insertional sequence-mediated gene deletion of a previously tdh-positive strain.Citation17,18 Finally, non-toxigenic V. parahaemolyticus isolates may possess novel virulence mechanisms.
The investigation of V. parahaemolyticus virulence mechanisms requires a suitable infection model. Several models have previously been used including orogastric and peritoneal mouse models, orogastric infection of infant rabbits and rabbit ileal-loop models. Oro-gastric models reveal diarrhoea and enteritis and have been useful for studying intestinal colonisation and pathogenesis. Galleria mellonella larvae of the greater wax moth have been investigated as an infection model since the 1980'sCitation19 but over the past few years there has been an escalation of interest in this model and the larvae have been shown to be susceptible to a wide range of bacterial and fungal pathogens.Citation20,21 In this study we investigated the utility of G. mellonella to identify virulence mechanisms of V. parahaemolyticus. This model has also allowed us to investigate mechanisms of virulence of non-toxigenic V. parahaemolyticus isolates, providing new insight into their clinical significance.
Results
The presence of non-toxigenic strains from clinical samples
We examined 10 strains of V. parahaemolyticus, 2 originating from the environment while the rest were clinical samples, these have all been listed in . Using PCR, we screened 6 clinical V. parahaemolyticus isolates previously known to be negative for virulence genes (tdh−/trh−/ T3SS2−) () as well as 2 environmental strains for the species marker toxR gene. All of the V. parahaemolyticus isolates were positive for the species marker toxR. We then confirmed that 7 of the V. parahaemolyticus strains were negative for the tdh, trh and T3SS2 genes. These included V. parahaemolyticus PSU 3384 and PSU 3565, isolated from patients in Thailand, in 2005.Citation13 and T03, T08, T023 and T024 isolated from patients in Italy, in 2010.Citation15 We also found one environmental isolate V13/003A from the UK which lacked these genes. The environmental strain V05/70, isolated from shellfish in Portugal, was positive for the trh and T3SS2β genes. The sequenced reference strain, V. parahaemolyticus RIMD 2210633,Citation22 was positive for the tdh and T3SS2α genes. Clinical strain V. parahaemolyticus G35, isolated from a recent outbreak in Spain in 2012 and serotyped as O4:K12,Citation23 was positive for tdh, trh, T3SS2α and T3SS2β genes.
Table 1. Vibrio parahaemolyticus strains used in this study.
V. parahaemolyticus shows virulence in G. mellonella model
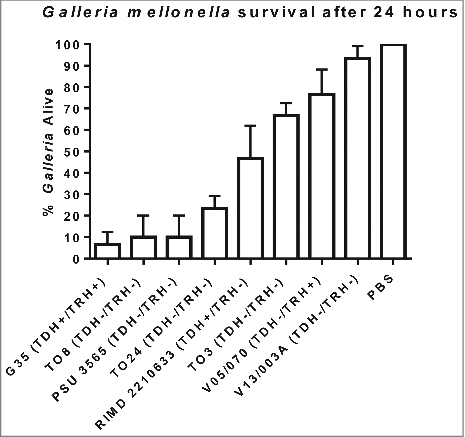
We next challenged groups of 10 G. mellonella larvae by injection into the foreleg with 100 CFU of V. parahaemolyticus G35, T08, T03, T024, PSU 3565, V05/70, V13/003A or RIMD 2210633 and the results are shown in . Approximately 20% of the larvae survived 24 h after inoculation with strains G35, T08, PSU 3565 and T024 (Statistically significant when compared to PBS using one way-ANOVA P < 0.0001). The O4:K12 serotype, which until 2012 was unique to the North West pacific region of the USA, has been shown to be the more virulent in humans resulting in a higher attack rate during outbreaks compared to other pathogenic strains.Citation23 In 2012, this serotype O4:K12 was responsible for an outbreak in Spain where 51 cases were detected among the 65 people who were exposed to the pathogen which represents a 78% attack rate. We found that a serotype O4:K12 isolate (G35) was the most virulent strain in G. mellonella and by 24 h post challenge less than 7% of the larvae survived (). The non-toxigenic clinical isolates (T08, T024 and PSU 3565) which lacked the tdh, trh and T3SS2 genes were virulent in G. mellonella with less than 24% survival of larvae at 24 h post challenge ((P < 0.0001). In contrast the non-toxigenic environmental isolate V13/003A, caused only 6% mortality of G. mellonella larvae after 24 h as seen in (not statistically significant). The LD50 was not determined for each of these strains but instead equivalent number of bacterial cells were used to compare their virulence in G. mellonella. We also found that clinical strain V. parahaemolyticus T03 which was non-toxigenic showed almost 65% survival rate after 24 h in the larvae model. Whilst we were unable to carry out genome sequencing on strain T03 (due to completion of project) we obtained a positive PCR result when using PCR primers designed to detect mutT (data not shown) in strain T03. When we tested V. parahaemolyticus strain RIMD2210633, a well characterised clinical TDH positive reference strain that has had its genome fully sequenced, the larvae showed 47% survival (). We also tested for comparison V05/070 an environmental TRH positive strain which showed 77% survival in the insect model.
Figure 1. Survival of G. mellonella following infection with 100 CFU per larvae of V. parahaemolyticus strains RIMD 2210633, G35, T08, PSU 3565, T024, T03, V05/070 or V13/003A. The results shown are the means of three experiments, each using groups of 10 larvae per strain. The error bars indicate standard deviation.
Multi locus sequence analysis (MLSA)
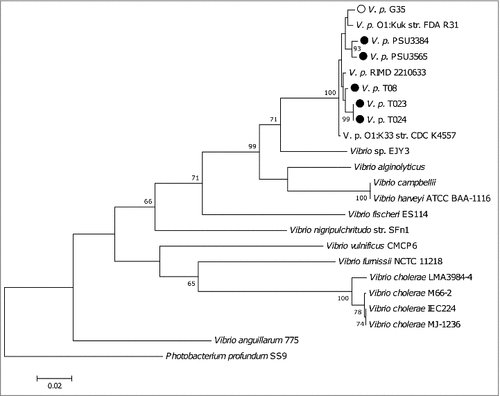
We genome sequenced 5 non-toxigenic clinical V. parahaemolyticus strains () (T08, T023, T024, PSU 3565 and PSU 3384) and one toxigenic strain (G35). MLSA, using the recA, gyrB, pyrH and atpA sequences, revealed that all of the V. parahaemolyticus strains grouped together and were distinct from closely related bacterium Photobacterium profundum (). MLSA revealed that strains T023 and T024 shared more than 99% similarity and are likely clones.
Identification of functional insertional sequences (ISVpa3)
To determine whether the clinical non-toxigenic strains T08, T023, T024, PSU 3384 and PSU 3565 were derived from previously toxigenic strains harbouring tdh/trh and genes for T3SS2 we searched for functional insertional sequences (ISVpa3) that surrounded the PaI-7 described by Kamruzzaman, et al. (2008) using BLASTn. ISVpa3 region flanks the two tdh sequences that can be found in tdh+ strains. We found that strain G35, had a sequence with 98% match to ISVpa3, as expected for a tdh and trh positive strain. Strains PSU 3384, T023 and T024 possessed DNA sequences with 43% matches to the ISVpa3. These results suggests that it is unlikely that tdh/trh and the genes for T3SS2 had been present in strains prior to infection. Strains T08 and PSU 3565 did not possess the ISVpa3 sequence.
Confirming the presence/absence of classical virulence factors
We confirmed the presence or absence of known virulence factors in the genome sequences of 5 non-toxigenic V. parahaemolyticus strains T08, T023, T024, PSU 3565 and PSU 3384 and one toxigenic strain G35 by carrying out BLASTn searches. shows a summary of the main virulence factors and pathogenicity islands present in Reference strain RIMD2210633 and their corresponding absence/presence in the 6 genome-sequenced strains. T3SS1 was found to be present in all strains. We found that VPaI-7 on which the genes for TDH, TRH and the T3SS2 are localised was absent in the five non-toxigenic strains we sequenced. Also absent was VPaI-1, 4, 5 and 6 from the five strains while VPaI-2 and 3 are partially present in the non-toxigenic strains.
Identification of a MutT/Nudix family protein
In order to identify genes unique to the clinical non-toxigenic strains we compared their genomes to available genome sequences of clinical toxigenic strains. We used Rapid Annotations using Subsystem Technology (RAST) to identify genes and found mutT, encoding a predicted MutT/Nudix family protein, in all five non-toxigenic strains. The predicted MutT/Nudix family protein, in all five non-toxigenic V. parahaemolyticus strains shared homology with nudix hydrolases from many different bacteria including Bacillus anthracis, Legionella pneophillia and Lactobacillus casei. The gene nudA encoding a Nudix hydrolase was shown to be an important virulence factor involved in resisting stress in Legionella pneumophila.Citation24 Using PCR, we confirmed the presence of mutT in the V. parahaemolyticus strains that we genome sequenced. The mutT gene was absent in the environmental strains V13/003A and V05/070 that showed low virulence in G. mellonella ().
A mutT mutant shows impaired virulence in G. mellonella
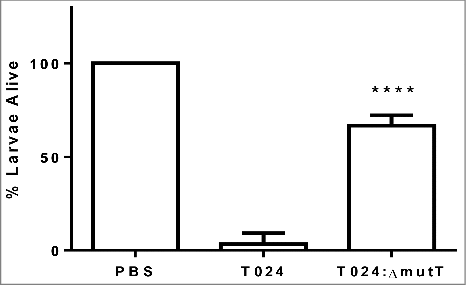
To determine if mutT was involved in virulence in V. parahaemolyticus we created an in frame deletion mutant in the strain T024, called T024:ΔmutT. Groups of 10 G. mellonella larvae were challenged by injection into the foreleg with 1–10 CFU of V. parahaemolyticus T024 or T024:ΔmutT (). Survival at 48 h post challenge was observed. Following challenge with V. parahaemolyticus T024:ΔmutT, 67% of infected larvae survived compared to 3% of larvae infected with wild-type (P < 0.0001). These data demonstrates that mutT plays a role in virulence in G. mellonella.
A mutT mutant shows reduced colony size
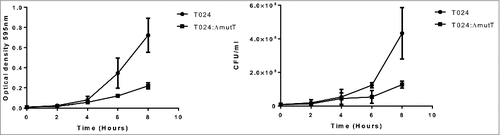
The deletion mutant of nudA in L. pneumophila grew slowly in liquid media and on solid media and had a smaller colony size than its parent strain.Citation24 In this study we chose minimal media which contained minimal nutrients possible to assess growth and colony size of the wild type and mutant strains. Wild type and mutant strains were preadapted to the minimal media by growing overnight in minimal media and then this was used to inoculate fresh media with equal numbers of the parent or mutant and incubated at 30°C. The wildtype strain T024 had a higher OD than the T024:ΔmutT at 8 h, while the CFU counts were similar (). Although we did not examine the cells microscopically V. parahaemolyticus T024:ΔmutT grew smaller in colony size than the T024 wild type strain on agar.
Figure 3. Growth of T024 or T024:ΔmutT at 30°C in minimal media measured by optical density (left panel) or CFU (right panel). At 8 hr there is significant difference between the wildtype and the T024:ΔmutT (determined by a paired T test P < 0.05) while no significant difference is seen with the cell counts.
Discussion
In this study we report for this first time that G. mellonella is susceptible to infection with V. parahaemolyticus. Previous studies have used G. mellonella to identify virulence associated genes in a range of pathogens including Pseudomonas aeruginosa.Citation25 Enterococcus faecalis,Citation26 Staphylococcus aureusCitation27 Yersinia pseudotuberculosisCitation28 and Campylobacter jejuni.Citation29 G. mellonella possess haemocytes that share a high degree of structural and functional similarity with mammalian phagocytic cells. The cellular response of G. mellonella to infection involves haemocytes that ingest bacteria and generate bactericidal products such as superoxide via a respiratory burst.Citation30 Haemocyte nodulation results in melanisation through the action of phenoloxidase.Citation31,32 The degree of melanisation in infected larvae results in a mottled grey to uniform black appearance, depending on the infecting bacteria. Insect larvae can be infected at 37°C, ensuring expression of temperature regulated virulence genes. Moreover, a defined infection site and the ability to challenge larvae with exact doses allows the virulence of strains to be compared. Our study shows that virulence in G. mellonella appears to correlate with the ability of isolates to cause disease in humans and demonstrates the value of G. mellonella for the identification of genes associated with virulence in V. parahaemolyticus. Current infection models to study V. parahaemolyticus infections include mammals such as mouse and infant rabbits. Although these mammalian models allow the study of virulence mechanisms, they are expensive, require specialist training, and need ethical approval before studies can be carried out. The use of a non-mammalian model to study V. parahaemolyticus such as larvae allows high throughput screening of a large number of V. parahaemolyticus strains at low costs. In this study we used G. mellonella as an initial screen to determine virulence of V. parahaemolyticus and reduce the reliance on experimental mammals. The availability of this infection model will be of value to others investigating mechanisms of virulence of V. parahaemolyticus. Furthermore, the simplicity of this insect model over other non-mammalian models will help the scientific community to better understand and investigate pathogenic traits of V. parahaemolyticus strains.
Previous work has attributed the ability of V. parahaemolyticus to cause disease in humans to the presence of genes encoding the TDH and/or TRH haemolysins and to the T3SS2 genes. Environmental strains of V. parahaemolyticus carry these virulence genes less frequently and are thought not able to illicit disease in humans.Citation33 However, there is growing evidence that strains lacking these genes can cause disease. For example, at Hat Yai Hospital in South Thailand 10% of V. parahaemolyticus infections are attributed to non-toxigenic strains. We showed in this study that a set of non-toxigenic V. parahaemolyticus strains were phylogenetically similar to other V. parahaemolyticus strains and did not carry the conventional virulence genes associated with disease such as tdh/trh and the genes for the T3SS2. We have shown that G. mellonella (wax moth) larvae are susceptible to a lethal infection with toxigenic and non- toxigenic clinical strains of V. parahaemolyticus. However, we noted some important differences in the pathogenesis of disease. Signs of infection after challenge with toxigenic strains occurred within a short period of time (<2 h) and included melanisation, reduced motility/movement, faecal staining and death. When clinical non-toxigenic V. parahaemolyticus strains were injected into larvae at comparable doses they caused disease and death in the larvae, but the rapid onset of melanisation was not observed. These findings suggest that the rapid effects seen after dosing with toxigenic strains may be a consequence of the direct action of the TDH or TRH toxins on the larvae.
Nudix hydrolases have been shown to be involved in virulence of a number of different pathogens.Citation24,34,35 For example, in Legionella pneumophila NudA is an important stress prevention factor and nudA mutants showed they were outcompeted in macrophage studies by fourfold by the parent in competition studies.Citation24 Further competition studies in guinea pigs also showed that the nudA mutant of Legionella pneumophila was outcompeted by its parent in both lung and spleen.Citation24 In this study we identified mutT, a nudix family protein present in the non-toxigenic strains of V. parahaemolyticus and suggest it may have a role in establishment of disease in strain T024. The reduced colony size observed of T024:ΔmutT did not lead to an altered growth rate. We were unable to construct a complemented strain of mutT in T024 to show that there were no polar effects that may have occurred from genetic manipulation in the T024 mutT mutant. However, during our initial screening of mutT mutants we found that multiple colonies gave comparable results in growth curves and initial screening in the G. mellonella model. Further studies where mutT mutants constructed in strains PSU3565 and T08 may help support that mutT is a general virulence factor in V. parahaemolyticus species. Nudix hydrolases are able to hydrolyse organic pyrophosphates, such as nucleoside diphosphates and triphosphates, as well as nucleotide sugars. Nudix hydrolase substrates or alarmones are hypothesised to be produced in high concentration in response to heat shock or oxidative stress. Further investigation is required to establish the function and role of mutT in V. parahaemolyticus strains that harbour this gene and the role it may play in establishing disease. Analysis of freely available complete genomes on NCBI (N = 17) found the mutT gene to be present in 3 environmental strains of V. parahaemolyticus (FORC 006, FORC 023 and FORC 022) that did not harbour any genes for tdh/trh and T3SS2. Thus, it may be important to determine the distribution of this gene in a broader range of clinical and environmental isolates of V. parahaemolyticus, to establish the role of mutT in virulence and to establish whether this gene is a useful marker of non-toxigenic clinical strains of V. parahaemolyticus.
Methods and materials
Strains and cultures used
Bacterial strains used in this study are shown in . V. parahaemolyticus strains were initially cultured aerobically onto selective media Thiosulphate Citrate Bile Sucrose (TCBS) agar (Oxoid) at 37°C for 24 h to check for contamination. For enumeration of colony counts we used Marine Agar (Conda lab, India) at 30°C for 24 h and for routine subculturing and growth Luria-Bertani (LB) agar at 37°C for 18 h was used. For growth curves, wildtype T024 and T024:ΔmutT were grown in minimal media with aeration at 30°C and optical readings and colony counts were carried out on Marine agar at 0, 2, 4, 6 and 8 h. For virulence assays the V. parahaemolyticus strains were grown in Marine Broth (Conda Lab, India) at 37°C.
PCR identification of genes
Bacterial cells were harvested and genomic DNA was extracted using Wizard Genomic DNA Purification Kit (Promega A1125). A ToxR targeted PCR () was used to confirm the identification of V. parahaemolyticus using primers described previously.Citation36 The tdh and trh genes were detected using previously described primers ().Citation37 Multiplex PCRs were carried out to detect the T3SS1 and T3SS2α genes using primers described previously by Noriea et al.Citation16 while T3SS2β genes were detected using primers described previously by Okada et al.Citation38 PCR was used to confirm the presence of the mutT gene in V. parahaemolyticus using the primers 5′-ATCCCCGGCGTTGCGGGTGTCATT-3′ and 5′-GCCAACGAAGTACGCTGAATCATTTTC-3′. The PCR amplification cycle for amplification of mutT consisted of 15 minutes at 96°C, followed by 30 cycles of 1 minute at 94°C, 1.5 minutes at 60.5°C and 1.5 minutes at 72°C, and finally with a single extension time of 7 minutes at 72°C. All PCR reactions described above were carried out using Hot Start Taq (Qiagen) and set up following the manufactures instructions. PCR reactions were run on a 1.5% agarose gel stained with SYBR Safe DNA stain and visualised using Chemidoc imaging system equipped with QuantityOne software (BioRad).
Table 2. Oligonucleotide primers used in this study.
Table 3. Presence or absence of chromosomal regions in 5 non-toxigenic strains (T08, T023, T024, PSU 3565 and PSU 3384) and 1 toxigenic strain (G35) when compared to reference strain RIMD2210633 and their corresponding absence/presence in the 6 genome-sequenced strains.
Infection of galleria mellonella larvae
G. mellonella larvae were purchased from Livefoods UK Ltd (Rooks Bridge, Somerset, UK) or TruLarv™ (Biosystems Technology, Exeter, Devon, UK).Citation28,39 Larvae weighing between 0.2 – 0.3 g were chosen for experiments. For each experiment a total of 10 larvae were used per strain to be tested. The larvae were infected by micro-injection (Hamilton Ltd) into the right foremost proleg with 100 CFU of V. parahaemolyticus in 10 µl volume which had been grown in Marine Broth at 37°C and washed twice in PBS. Bacterial cell counts were carried out by plating serial dilutions of the inoculum onto Marine agar. For control purposes 10 larvae were inoculated with PBS and a further 10 were left uninoculated. The larvae were incubated at 37°C and survival was recorded for all strains after 24 h. An additional 48 h time point was needed for experiment with the T024:ΔmutT. Larvae were scored as dead when they ceased moving, and failed to respond when gently manipulated with a pipette tip. Observation findings were also recorded if larvae colour changed from their normal pale cream coloration to brown or black indicative of melanisation.
Genome sequencing, assembly and annotation
Seven V. parahaemolyticus strains () were genome sequenced. Genomic DNA (gDNA) was extracted using the Wizard Genomic DNA Purification Kit (Promega). Library prep was carried out by Exeter DNA sequencing service. In brief, DNA was concentrated using GeneRead kit (Lot No. 145025210) and end repair and adenylation of fragments was carried using NEXTflex Rapid DNAseq kit (#5144–02) according to manufactures instructions. Purification and concentration of PCR amplified library was carried out according to GeneRead kit instructions. Genome wide sequence data was generated using an Illumina Hiseq 2500 platform. Raw sequencing reads were screened for contamination and PCR duplicates and quality filtered using fastq-mcf.Citation48 This filtered dataset was then assembled de-novo using velvet and velvet optimiserCitation40 (final hash lengths and coverage cut offs can be found in Supplementary Data ). De-novo assemblies were annotated using Rapid Annotation using Subsystems Technology (RAST).Citation41,42 Assembly metrics were generated using the Quast package.Citation43
Comparative genomics
To assess the presence or absence of genes within each of the strains, the sequencing reads were aligned against a “pan genome” consisting of all the genes annotated in each of the experimental strains. The gene sequences were clustered using UCLUSTCitation44 and one sequence was selected (by length) from each orthologous cluster. Trimmed sequencing reads were then aligned to this “pan-genome” using BOWTIE2Citation45 to assess the presence of each gene in each strain. The benefit of this approach is that it reduces the effect of poor assemblies on the comparative genomic analysis. It is only required for each gene to have been assembled once for it to appear in the “pan-genome” and aligning sequencing reads means even if the gene was not assembled correctly in any of the other strains, the reads will align to the target sequence from any of those strains in which it appears.
Multi locus sequence analysis (MLSA)
Multi locus sequence analysis (MLSA) was conducted on V. parahaemolyticus sequenced strains and genomes in the NCBI genomes database (July 2017). The MLSA was conducted using 4 conserved housekeeping genes (gryB, pyrH, recA, atpA). These genes were extracted from the genome sequences using blast and in house scripts to parse and extract results. The MLSA sequences were aligned using MUSCLECitation46 and data was converted into a phylogenetic tree using MEGA 7Citation47 with a bootstrap value of 100 and using Photobacterium profundum as the root of the tree.
mutT mutant construction
DNA fragments (500bp) including upstream and downstream regions of mutT and flanked by SacI and SphI restriction enzymes were created using PCR and ligation. The DNA fragment was cloned into plasmid pDM4 via the SacI and SphI sites. The presence of the cloned DNA was confirmed by PCR using primers described in (mutT flank 1 and 2). The plasmid pDM4-mutT was maintained in E. coli GT115 cells and selected on LB agar containing 50 µg/ml chloramphenicol.
Conjugation of V. parahaemolyticus T024
One ml of an overnight culture of the recombinant E.coli GT115 pdm4-mutT (donor strain), E.coli pKR2013 (helper strain) and the V. parahaemolyticus T024 (recipient strain) were centrifuged for 2 minutes. Supernatants were discarded and the pellets re-suspended in 0.5 ml LB medium. A 10 µl aliquot was pipetted onto a sterile nitrocellulose membrane on LB agar plate, either individually or with donor, helper and recipient mixed together (ratio 1:1:4), and incubated overnight at 37°C. The cells were then re-suspended in 1ml sterile PBS. Aliquots of 100 µl were plated onto LB agar plates supplemented with 100 µg/ml chloramphenicol and incubated overnight at 37ºC. Colony growth was scraped off using a sterile 10 µl loop and re-suspending in 1ml of sterile PBS and aliquots of 100 µl were plated onto TCBS plate's supplemented with 50 µg/ml chloramphenicol. After incubation at 37ºC for 5–7 days, colonies were transferred onto fresh LB plates containing chloramphenicol. The transconjugants were grown in LB broth without supplementation overnight, serially diluted in PBS and plated onto salt free LB agar containing 10% (w/v) sucrose. The plates were incubated at 24ºC for 2 to 5 days and colonies screened for chloramphenicol sensitivity and on TCBS agar. In order to confirm that chloramphenicol sensitive colonies contained the desired mutation, PCR was carried out using ΔMutT confirmation primers (). The deletion mutant (T024:ΔmutT) was used in our subsequent experiments.
Statistical analyses
Analysis was carried out on the differences between average survivals of larvae when infected with different strains of V. parahaemolyticus. These values were tested for significance by performing a one way ANOVA comparing each of the strains to PBS and were performed using the GraphPad Prism software (GraphPad Software, San Diego California USA).
KVIR_S_1384895.zip
Download Zip (10.4 KB)Acknowledgements
We acknowledge Dr Konrad Paszkiewicz, Dr Karen Moore, Exeter DNA Sequencing Services and funding from Wellcome Trust Institutional Strategic Support Fund (WT097835MF), Wellcome Trust Multi User Equipment Award (WT101650MA) and Medical Research Council Clinical Infrastructure Funding (MR/M008924/1) for supporting whole genome sequencing of strains within this study. We also thank Florentina Papanicola and Nicole Needham for their help with gathering preliminary data to support this study.
Funding
None of the authors of this manuscript have any commercial or other association that might pose a conflict of interest. This work was partly supported by Wellcome Trust Institutional Strategic Support Fund (WT097835MF), Wellcome Trust Multi User Equipment Award (WT097835MF) Medical Research Council Clinical Infrastructure Funding (MR/M008924/1) and Biotechnology and Biological Sciences Research Council (BBSRC) funding (BB/N016513/1).
References
- Joseph SW, Colwell RR, Kaper JB. Vibrio parahaemolyticus and related halophilic vibrios. Crit Rev Microbiol. 1982;10:77-124. doi:10.3109/10408418209113506. PMID:6756788
- Letchumanan V, Chan KG, Lee LH. Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front Microbiol. 2014;5:705. doi:10.3389/fmicb.2014.00705. PMID:25566219
- Letchumanan V, Pusparajah P, Tan LT, Yin WF, Lee LH, Chan KG. Occurrence and antibiotic resistance of Vibrio parahaemolyticus from shellfish in selangor, Malaysia. Front Microbiol. 2015;6:1417. doi:10.3389/fmicb.2015.01417. PMID:26697003
- Letchumanan V, Yin WF, Lee LH, Chan KG. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front Microbiol. 2015;6:33. doi:10.3389/fmicb.2015.00033. PMID:25688239
- Krachler AM, Ham H, Orth K. Outer membrane adhesion factor multivalent adhesion molecule 7 initiates host cell binding during infection by gram-negative pathogens. Proc Natl Acad Sci U S A. 2011;108:11614-9. doi:10.1073/pnas.1102360108. PMID:21709226
- Trosky JE, Li Y, Mukherjee S, Keitany G, Ball H, Orth K. VopA inhibits ATP binding by acetylating the catalytic loop of MAPK kinases. J Biol Chem. 2007; 282:34299-305. doi:10.1074/jbc.M706970200. PMID:17881352
- Liverman AD, Cheng HC, Trosky JE, Leung DW, Yarbrough ML, Burdette DL, Rosen MK, Orth K. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc Natl Acad Sci U S A. 2007;104:17117-22. doi:10.1073/pnas.0703196104. PMID:17942696
- Kodama T, Rokuda M, Park KS, Matsuda S, Iida T, Honda T. Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system 2. Cell Microbiol. 2007;9:2598-609. doi:10.1111/j.1462-5822.2007.00980.x. PMID:17645751
- Hiyoshi H, Kodama T, Saito K, Gotoh K, Matsuda S, Akeda Y, Honda T, Iida T. VopV, an F-actin-binding type III secretion effector, is required for Vibrio parahaemolyticus-induced enterotoxicity. Cell Host Microbe. 2011;10:401-9. doi:10.1016/j.chom.2011.08.014. PMID:22018240
- Akeda Y, Kodama T, Saito K, Iida T, Oishi K, Honda T. Identification of the Vibrio parahaemolyticus type III secretion system 2-associated chaperone VocC for the T3SS2-specific effector VopC. FEMS Microbiol Lett. 2011;324:156-64. doi:10.1111/j.1574-6968.2011.02399.x. PMID:22092817
- Park KS, Ono T, Rokuda M, Jang MH, Okada K, Iida T, Honda T. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect Immun. 2004;72:6659-65. doi:10.1128/IAI.72.11.6659-6665.2004. PMID:15501799
- Broberg CA, Calder TJ, Orth K. Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 2011;13:992-1001. doi:10.1016/j.micinf.2011.06.013. PMID:21782964
- Wootipoom N, Bhoopong P, Pomwised R, Nishibuchi M, Ishibashi M, Vuddhakul V. A decrease in the proportion of infections by pandemic Vibrio parahaemolyticus in Hat Yai Hospital, southern Thailand. J Med Microbiol. 2007;56:1630-8. doi:10.1099/jmm.0.47439-0. PMID:18033832
- Thongjun J, Mittraparp-Arthorn P, Yingkajorn M, Kongreung J, Nishibuchi M, Vuddhakul V. The Trend of Vibrio parahaemolyticus Infections in Southern Thailand from 2006 to 2010. Tropical medicine and health. 2013;41:151-6. doi:10.2149/tmh.2013-06. PMID:24478592
- Ottaviani D, Leoni F, Serra R, Serracca L, Decastelli L, Rocchegiani E, Masini L, Canonico C, Talevi G, Carraturo A. Nontoxigenic Vibrio parahaemolyticus strains causing acute gastroenteritis. J Clin Microbiol. 2012;50:4141-3. doi:10.1128/JCM.01993-12. PMID:23052317
- Noriea NF, 3rd, Johnson CN, Griffitt KJ, Grimes DJ. Distribution of type III secretion systems in Vibrio parahaemolyticus from the northern Gulf of Mexico. J Appl Microbiol. 2010;109:953-62. doi:10.1111/j.1365-2672.2010.04722.x. PMID:20408916
- Bhoopong P, Palittapongarnpim P, Pomwised R, Kiatkittipong A, Kamruzzaman M, Nakaguchi Y, Nishibuchi M, Ishibashi M, Vuddhakul V. Variability of properties of Vibrio parahaemolyticus strains isolated from individual patients. J Clin Microbiol. 2007;45:1544-50. doi:10.1128/JCM.02371-06. PMID:17344357
- Kamruzzaman M, Bhoopong P, Vuddhakul V, Nishibuchi M. Detection of a functional insertion sequence responsible for deletion of the thermostable direct hemolysin gene (tdh) in Vibrio parahaemolyticus. Gene. 2008;421:67-73. doi:10.1016/j.gene.2008.06.009. PMID:18598741
- Morton DB, Dunphy GB, Chadwick JS. Reactions of hemocytes of immune and non-immune Galleria mellonella larvae to Proteus mirabilis. Developmental and comparative immunology. 1987;11:47-55. doi:10.1016/0145-305X(87)90007-3. PMID:3109970
- Tsai CJ, Loh JM, Proft T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. 2016;7:214-29. doi:10.1080/21505594.2015.1135289. PMID:26730990
- Champion OL, Wagley S, Titball RW. Galleria mellonella as a model host for microbiological and toxin research. Virulence. 2016;7(7):840-5.
- Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet. 2003;361:743-9. doi:10.1016/S0140-6736(03)12659-1. PMID:12620739
- Martinez-Urtaza J, Baker-Austin C, Jones JL, Newton AE, Gonzalez-Aviles GD, DePaola A. Spread of Pacific Northwest Vibrio parahaemolyticus strain. N Engl J Med. 2013;369:1573-4. doi:10.1056/NEJMc1305535. PMID:24131194
- Edelstein PH, Hu B, Shinzato T, Edelstein MA, Xu W, Bessman MJ. Legionella pneumophila nuda is a nudix hydrolase and virulence factor. Infect Immun 2005;73:6567-76. doi:10.1128/IAI.73.10.6567-6576.2005. PMID:16177332
- Jander G, Rahme LG, Ausubel FM. Positive correlation between virulence of pseudomonas aeruginosa mutants in mice and insects. Journal of bacteriology 2000;182:3843-5. doi:10.1128/JB.182.13.3843-3845.2000. PMID:10851003
- Lebreton F, Riboulet-Bisson E, Serror P, Sanguinetti M, Posteraro B, Torelli R, Hartke A, Auffray Y, Giard JC. ace, Which encodes an adhesin in enterococcus faecalis, is regulated by ers and is involved in virulence. Infection and immunity. 2009;77:2832-9. doi:10.1128/IAI.01218-08. PMID:19433548
- Peleg AY, Monga D, Pillai S, Mylonakis E, Moellering RC, Jr., Eliopoulos GM. Reduced susceptibility to vancomycin influences pathogenicity in staphylococcus aureus infection. J Infect Dis. 2009;199:532-6. doi:10.1086/596511. PMID:19125671
- Champion OL, Cooper IA, James SL, Ford D, Karlyshev A, Wren BW, Duffield M, Oyston PC, Titball RW. Galleria mellonella as an alternative infection model for Yersinia pseudotuberculosis. Microbiology (Reading, England). 2009;155:1516-22. doi:10.1099/mic.0.026823-0. PMID:19383703
- Champion OL, Karlyshev AV, Senior NJ, Woodward M, La Ragione R, Howard SL, Wren BW, Titball RW. Insect Infection model for campylobacter jejuni reveals that o-methyl phosphoramidate has insecticidal Activity. The Journal of infectious diseases. 2010. doi:10.1086/650494. PMID:20113177
- Bergin D, Reeves EP, Renwick J, Wientjes FB, Kavanagh K. Superoxide production in Galleria mellonella hemocytes: identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect Immun. 2005;73:4161-70. doi:10.1128/IAI.73.7.4161-4170.2005. PMID:15972506
- Jiravanichpaisal P, Lee BL, Soderhall K. Cell-mediated immunity in arthropods: hematopoiesis, coagulation, melanization and opsonization. Immunobiology 2006;211:213-36. doi:10.1016/j.imbio.2005.10.015. PMID:16697916
- Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect biochemistry and molecular biology. 2002;32:1295-309. doi:10.1016/S0965-1748(02)00092-9. PMID:12225920
- Powell A, Baker-Austin C, Wagley S, Bayley A, Hartnell R. Isolation of pandemic Vibrio parahaemolyticus from UK water and shellfish produce. Microbial ecology. 2013;65:924-7. doi:10.1007/s00248-013-0201-8. PMID:23455432
- Badger JL, Wass CA, Kim KS. Identification of escherichia coli K1 genes contributing to human brain microvascular endothelial cell invasion by differential fluorescence induction. Mol Microbiol. 2000;36:174-82. doi:10.1046/j.1365-2958.2000.01840.x. PMID:10760174
- Conyers GB, Bessman MJ. The gene, ialA, associated with the invasion of human erythrocytes by Bartonella bacilliformis, designates a nudix hydrolase active on dinucleoside 5′-polyphosphates. J Biol Chem. 1999;274:1203-6. doi:10.1074/jbc.274.3.1203. PMID:9880487
- Kim YB, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol. 1999;37:1173-7. PMID:10074546
- Tada J, Ohashi T, Nishimura N, Shirasaki Y, Ozaki H, Fukushima S, Takano J, Nishibuchi M, Takeda Y. Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol Cell Probes. 1992;6:477-87. doi:10.1016/0890-8508(92)90044-X. PMID:1480187
- Okada N, Iida T, Park KS, Goto N, Yasunaga T, Hiyoshi H, Matsuda S, Kodama T, Honda T. Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect Immun. 2009;77:904-13. doi:10.1128/IAI.01184-08. PMID:19075025
- Wand ME, Muller CM, Titball RW, Michell SL. Macrophage and Galleria mellonella infection models reflect the virulence of naturally occurring isolates of B. pseudomallei, B. thailandensis and B. oklahomensis. BMC Microbiol. 2011;11:11. doi:10.1186/1471-2180-11-11. PMID:21241461
- Zerbino DR, Birney E. Velvet: Azlgorithms for de novo short read assembly using de bruijn graphs. Genome Res. 2008;18:821-9. doi:10.1101/gr.074492.107. PMID:18349386
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008;9:75. doi:10.1186/1471-2164-9-75. PMID:18261238
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42:D206-14. doi:10.1093/nar/gkt1226. PMID:24293654
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072-5. doi:10.1093/bioinformatics/btt086. PMID:23422339
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460-1. doi:10.1093/bioinformatics/btq461. PMID:20709691
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357-9. doi:10.1038/nmeth.1923. PMID:22388286
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792-7. doi:10.1093/nar/gkh340. PMID:15034147
- Kumar S, Stecher G, Tamura K. MEGA7: molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870-4. doi:10.1093/molbev/msw054. PMID:27004904
- Erik Aronesty 2011. Ea-utils: “Command-line tools for processing biological sequencing data”; https://github.com/ExpressionAnalysis/ea-utils