ABSTRACT
Candida species are a major cause of invasive fungal infections. While Candida albicans, C. glabrata, C. parapsilosis, and C. tropicalis are the most dominant species causing life-threatening candidiasis, C. auris recently emerged as a new species causing invasive infections with high rates of clinical treatment failures. To mimic initial phases of systemic Candida infections with dissemination via the bloodstream and to elucidate the pathogenic potential of C. auris, we used an ex vivo whole blood infection model. Similar to other clinically relevant Candida spp., C. auris is efficiently killed in human blood, but showed characteristic patterns of immune cell association, survival rates, and cytokine induction. Dual-species transcriptional profiling of C. auris-infected blood revealed a unique C. auris gene expression program during infection, while the host response proofed similar and conserved compared to other Candida species. C. auris-specific responses included adaptation and survival strategies, such as counteracting oxidative burst of immune cells, but also expression of potential virulence factors, (drug) transporters, and cell surface-associated genes. Despite comparable pathogenicity to other Candida species in our model, C. auris-specific transcriptional adaptations as well as its increased stress resistance and long-term environmental survival, likely contribute to the high risk of contamination and distribution in a nosocomial setting. Moreover, infections of neutrophils with pre-starved C. auris cells suggest that environmental preconditioning can have modulatory effects on the early host interaction. In summary, we present novel insights into C. auris pathogenicity, revealing adaptations to human blood and environmental niches distinctive from other Candida species.
Introduction
Candida species are among the most prevalent human fungal pathogens worldwide, and represent the fourth most common cause of nosocomial (hospital-acquired) bloodstream infections in the US [Citation1,Citation2]. C. albicans is the most common cause of candidiasis, and together with C. glabrata, C. parapsilosis, and C. tropicalis, accounts for around 90% of Candida bloodstream infections [Citation3–5]. Recently a new opportunistic Candida species, C. auris, has emerged and quickly spread to every permanently inhabited continent [Citation6]. This species is of clinical concern as most clinical isolates appear to be resistant against commonly used antifungal drugs, dramatically limiting therapeutic options [Citation7]. Numerous outbreaks in healthcare facilities have been reported in different regions worldwide, with C. auris bloodstream infection-associated mortality rates reaching around 30–60% [Citation8,Citation9]. C. auris rapidly emerged simultaneously in at least four different clades, each corresponding to a specific geographical region: South Asian clade I (representative strain: B8441 with reference genome available at the Candida Genome Database, CGD [Citation10]), East Asian clade II, African clade III (representative strain: B11221 with sequenced genome on CGD), and South American clade IV [Citation11–13]. The first clinically reported C. auris isolate from 2009 belongs to clade II and was derived from an unusual ear infection [Citation14,Citation15]. Recent C. auris infections and hospital outbreaks have been associated with strains from clades I, III, and IV [Citation16,Citation17]. In these cases, C. auris has been not only isolated from the skin of patients and medical devices, but was also recovered from blood, urine, peritoneal, vaginal, and respiratory samples [Citation9,Citation18]. While C. albicans and C. glabrata are usually found associated with the human host [Citation19–22], C. auris has been often isolated from the skin and the environment, including the hospital setting, suggesting that C. auris is well-adapted for survival outside the human body [Citation23–28]. The species closest related to C. auris is C. haemulonii, which has been isolated from environmental and superficial host niches [Citation9,Citation12,Citation29]. While C. haemulonii, similar to C. auris, shows a relatively high intrinsic resistance against antifungals, this related species lacks the ability to grow at elevated (host-relevant) temperatures. In contrast to C. haemulonii and most environmental fungi, C. auris has been shown to grow readily at 37°C, and even up to 42°C [Citation29]. Apart from thermal tolerance, its high salinity tolerance and resistance against other environmental stresses hint toward a remarkable ability of C. auris to survive different environmental conditions [Citation30,Citation31]. The virulence factors of C. auris are largely unknown or uncharacterized, but many orthologues of virulence-associated C. albicans genes can be found in the C. auris genome [Citation32]. For example, the chaperone Hsp90 is associated with C. albicans morphogenesis and virulence, and its C. auris orthologue was found to be involved in growth, morphology, and antifungal drug tolerance [Citation33]. Although different in vivo studies using Galleria [Citation34], Drosophila [Citation35], zebrafish [Citation36], or mice [Citation29,Citation37] clearly underline the ability of C. auris to cause invasive infection in animals, little is known about the pathogenicity mechanisms of this emerging fungus.
To disseminate throughout the human body, Candida cells have to enter the bloodstream where they face the immune system and nutrient limitations, especially for those micronutrients that are actively sequestered away by the hosts, such as iron [Citation38]. Multiple arms of the innate immune system, including the complement system, monocytes, and neutrophils immediately act against invading Candida cells in the bloodstream. Consequently, Candida species including C. albicans, C. glabrata, C. tropicalis, and C. parapsilosis developed survival strategies for confrontations with innate immune cells or components, especially human neutrophils [Citation39–46], monocytes or dendritic cells [Citation47,Citation48], and the complement system [Citation49–51]. One recent study described resistance of C. auris to killing by human neutrophils, associated with reduced recruitment and formation of neutrophil extracellular traps (NETs) compared to C. albicans [Citation36]. Other studies described the immune response of peripheral blood mononuclear cells (PBMC) to C. auris with regard to the pro-inflammatory response [Citation37,Citation52]. While most of these in vitro studies focused on one specific, isolated immune cell type, subsequent work has analyzed cellular and transcriptional responses of host and pathogens during bloodstream infections in a complex ex vivo whole blood infection model for C. albicans, C. glabrata, C. tropicalis, and C. parapsilosis [Citation53,Citation54]. In the present study, we use this ex vivo whole blood infection model to characterize and compare C. auris pathogenicity and its fungal adaptation and survival strategies which allow its subsequent systemic dissemination. Molecular and cellular events during bloodstream infection are characterized for C. auris in comparison to the other four investigated Candida spp., including transcriptional responses from the fungal and host sides using dual-species RNA sequencing. We use in vitro models to elaborate the crucial roles of human neutrophils during blood infection by C. auris. Fungal survival under conditions mimicking hospital settings is assayed for C. auris in comparison with other Candida species, as this seems to be a prerequisite for nosocomial spreading. Furthermore, we explore whether environmental conditions may prime C. auris for survival in the host.
In summary, our data suggest novel C. auris features that mediate environmental adaptation, host survival, and pathogenicity, which are in part unique or otherwise similar to other Candida species. These analyses enable us to integrate C. auris into the pathogenic landscape of clinically relevant Candida species.
Material & methods
Candida strains and cultivation
The Candida auris strains used in this study have been kindly provided by the Mycotic Diseases Branch, Centers for Disease Control and Prevention (CDC; Atlanta [Citation11]) and C. auris 1715 by Neil Gow (University of Exeter [Citation30]. In order to compare the properties of C. auris to other Candida species on the molecular level, we focused on the C. auris genome reference strain B8441, representing the South Asian clade I [Citation11,Citation12]. C. albicans strain SC5314 [Citation55], C. glabrata ATCC2001, C. tropicalis DSM4959, C. parapsilosis GA1 [Citation56], and C. haemulonii CBS 5149 T were used for comparison. These strains were also used in previous studies by Kämmer et al. and Pekmezovic et al. for transcriptional profiling [Citation53,Citation57].
Candida strains were routinely grown and maintained on YPD agar plates (1% yeast extract, 2% peptone, 2% glucose, 2% agar) at 30°C. For use in experiments, Candida cells were cultured overnight in liquid YPD (1% yeast extract, 2% peptone, 2% glucose) at 30°C, shaking at 180 rpm. Cells from overnight cultures were harvested by centrifugation, washed with PBS, and the cell number was adjusted to the desired concentration as indicated for each experiment.
Analysis of fungal growth
Fungal growth was analyzed in 96-well plates in YPD and other media as mentioned. Candida strains were added at a final density of 2 × 106 cells/well, and growth was determined by measuring the absorbance at 600 nm every 30 min for the indicated time period and temperature in a microplate reader (Tecan). Preparation of mouse organ homogenates was done according to Dunker et al. with stepwise filtering of homogenized liver or kidney from male C57BL/6 J mice for final organ concentrations of 0.025 g organs/ml [Citation58].
Ex vivo whole blood infection model
Ethics approval and consent to participate
Human peripheral blood was collected from healthy volunteers with written, informed consent. This study was conducted according to the principles expressed in the Declaration of Helsinki. The blood donation protocol and use of blood for this study were approved by the institutional ethics committee of the University Hospital Jena (permission number 2207–01/08).
Quantification of fungal survival
Human whole blood was freshly drawn from healthy volunteers and anticoagulated with recombinant Hirudin (Sarstedt). Candida cells were harvested as described in `Candida strains and cultivation`. Candida cells diluted in PBS (Phosphate buffered saline) were added to the blood at a concentration of 1 × 106 cells per ml and further incubated at 37°C. Samples were taken at indicated time points and plated on YPD in appropriate dilutions (in PBS) to determine colony forming units (CFUs) as a measure of fungal survival. These plates were incubated at 30°C to prevent side effects (cell-cell aggregations) due to C. albicans filamentation. Blood smears of Candida-infected samples were prepared at indicated time points and stained with May-Grünwald-Giemsa staining, dried and visualized microscopically for histological analysis.
Determination of immune cell association
Fungal cells from overnight cultures were stained with FITC (fluorescein isothiocyanate), added to freshly drawn human blood at a concentration of 1 × 106 cells per ml and incubated at 37°C. As in previous studies [Citation53,Citation59], samples from infected whole blood were taken at the indicated time points, specific conjugated antibodies were added (see below), treated with FACS Lysing Solution (BD), washed, and immediately analyzed with the BD FACSCanto II flow cytometer counting 100,000 events per sample. The gating strategy is based on plotting forward (FSC) and side scatter (SSC) to exclude cellular debris, followed by gating for single cells using FSC-W and FSC-H. The population of single cells was further plotted by using SSC-A and CD445 to get the population of leukocytes as well as by using FITC-A and SSC-A to get leukocytes associated with fungal cells. To distinguish different immune cells in both of these leukocyte populations, specific staining with the following conjugated antibodies was used CD45-PE-Cy7 (clone HI30, leukocytes), mouse anti-human CD3-PerCP (clone SK7, T cells), CD19-PE (clone HIB19, B cells), CD56-V450 (clone B159, NK cells), and CD66b-PE (clone G10F5, PMN) obtained from BioLegend. Monocytes were stained with mouse anti-human CD14-PerCP (clone 47–3D6) from Abcam. The presence of activation markers was determined with mouse anti-human CD11b-V450 (clone ICRF44) from BD and CD16-BV510 (clone 3G8), CD69-APC (clone FN50) from BioLegend. For raw data analysis, the software FlowJo v10.0.8 was used.
RNA isolation
At indicated time points, infected blood samples were split for the separate isolation of fungal and human RNA. To isolate human RNA, blood aliquots were added to PAXgene Blood RNA Tubes (PreAnalytiX) and processed with the PAXgene Blood RNA Kit (PreAnalytiX) according to the manufacturer’s instructions. To isolate fungal RNA, aliquots of infected blood were added to ice-cold water to lyse human cells, centrifuged, and immediately frozen in liquid nitrogen. The (fungal) cell pellet was further processed with the RiboPure-Yeast Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. RNA quantity was determined with a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific), and RNA quality was verified with the help of an Agilent 2100 Bioanalyzer (Agilent Technologies). Fungal and human RNA samples were pooled subsequently in a quantitative ratio of 1:6 taking different genome sizes into account. All samples were prepared in three biological replicates with independent donors.
RNA sequencing
Library preparation and RNA sequencing were carried out at Eurofins Genomics GmbH (Ebersberg, Germany) using the Illumina HiSeq 2500 platform. After poly(A) enrichment, mRNA was fragmented, and cDNA libraries were generated for each sample.
All preprocessing was done using the GEO2RNAseq pipeline (v0.100.1 [Citation60]). The raw reads were quality trimmed with Trimmomatic v0.39, rRNA depleted with SortMeRNA v2.1b and quality controlled with FastQC v0.11.8. The Homo sapiens genome GRCh38.p13 (release 100.38) and annotation were downloaded from the ENSEMBL database. C. albicans SC5314 assembly 22 (s07-m01-r100), C. auris B8441 (s01-m01-r05), C. glabrata CBS138 (s02-m07-r43), C. parapsilosis CDC317 (s01-m03-r41), and C. tropicalis MYA-3404 (version from 11 December 2013) genomes and corresponding genome annotations were downloaded from the Candida Genome Database (CGD). Sequencing reads were mapped against concatenated genomes comprised of H. sapiens and each Candida species using HiSat2 v2.1.0. For H. sapiens with C. albicans all reads that mapped equally well to both species were removed. This was necessary for correct downstream analysis with the diploid C. albicans reference genome. Transcriptome coverage was calculated as mapped reads multiplied by read length and divided by transcriptome length. featureCounts v1.34.0 was applied to count the number of uniquely mapped reads within annotated genes. For C. albicans with its diploid reference genome we counted uniquely and multimapped reads and merged counts across alleles by averaging. Human and pathogen genes were tested individually for significant differential expression. For further analysis we normalized all transcriptome data by applying Median Ratio Normalization (MRN) for each species separately. For each individual Candida species, we computed the log2(fold change) of each time point against the 0 mpi reference time point and thus normalized to species-specific base transcript levels. DESeq2 was used to calculate adjusted p-values based on count values. Afterward the following cutoffs were applied: adjusted p-value ≤ 0.01, abs(log2FC) ≥ 1.5 and MRN ≥ 1 for at least one time point. The generated RNA sequencing dataset analyzed in the present study has been deposited in NCBI’s Gene Expression Omnibus [Citation61] under the GEO record GSE179000. The RNA-seq data set for C. albicans, C. glabrata, C. tropicalis, and C. parapsilosis, originally generated in the study by Kämmer et al., had been deposited in NCBI’s Gene Expression Omnibus under the GEO record GSE114180 [Citation53].
Gene expression data analyses
The PCA biplot of MRN values of H. sapiens is based on all genes, and the PCA biplot of MRN values for the combined Candida species is based on all genes that are orthologous among all five Candida species. The PCA biplot of log2FC values of H. sapiens is based on all H. sapiens genes with an adjusted p-value ≤0.01 in at least one comparison, the one for the combined Candida species is based on all genes that are orthologues among all five Candida species and have an adjusted p-value ≤0.01 in at least one comparison. Orthologous genes between C. albicans, C. auris, C. glabrata, and C. parapsilosis and additionally to S. cerevisiae were mapped with orthology information from CGD. Since orthology information for C. tropicalis to the other four Candida spp. was not available in CGD, information for orthologous genes between C. tropicalis and S. cerevisiae was retrieved from the Candida Gene Order Browser (CGOB). Orthologous genes of C. tropicalis to the other four Candida spp. were subsequently mapped via identical gene names for S. cerevisiae.
Gene set enrichment analysis (GSEA 4.1.0 [Citation62]) was performed on the human MRN data sets (diff_of_classes method based on log2FC values vs 0 mpi), after conversion to Entrez identifier based on information downloaded from Ensembl Biomart [Citation63], using the Biocarta gene sets of the MSigDB (C2). Gene sets with a false discovery rate q < 0.05 were considered significantly enriched, and their normalized enrichment scores (NES) were plotted for all species and time points. Plots were generated using R 4.1.2 [Citation64] with ggplot2 [Citation65].
Quantification of cytokine levels
Infected blood samples taken at indicated time points were centrifuged to obtain plasma and immediately frozen in liquid nitrogen. Concentrations of the cytokines IL-1β, IL-6, IL-8, and TNF-α were determined by ELISA kits according to the manufacturer’s instructions (eBioscience). Cytokine levels were calculated from standard dilutions of the respective recombinant cytokines.
Infection of neutrophil granulocytes
Preparation of neutrophils and serum from human whole blood
Neutrophils were isolated from freshly drawn, whole blood of healthy volunteers using a gradient centrifugation method. Briefly, 1:1 PBS-diluted blood was layered on top of Lymphocyte separation medium (Capricorn) with Ficoll density gradient and centrifuged for 30 min at 700 g. After this, the polymorphonuclear cell (PMN) fraction on top of the erythrocytes was kept, while plasma and the other (cell) layers were removed. Residual erythrocytes were removed by 10–15 min incubation in a hypotonic lysing buffer on ice with subsequent centrifugation for 10 min 1400 rpm at 4°C. This was followed by two analogous washing steps with cold PBS. The resulting cell pellets were resuspended in RPMI1640 w/o phenol red + 10% autologous human serum at a concentration of 5 × 105 cells/ml. For the autologous human serum, blood from the same donors as the PMN isolation was drawn in serum tubes (Sarstedt) and kept for at least 1 h at RT. Subsequently, tubes were centrifuged for 10 min at 2500 g, and the serum was collected for each donor.
ROS assay
Neutrophil activation upon stimulation with Candida cells was determined by an oxidative burst assay, using the luminol-enhanced chemiluminescence method for the quantification of total reactive oxygen species (ROS). Freshly isolated neutrophils (see above) were seeded at a concentration of 5 × 104 cells/well in a 96 well plate (white, clear bottom, Corning) and incubated for about 1 h at 37°C 5% CO2, to let them attach. Candida cells, opsonized with human serum for 30 min at 37°C, were washed, and resuspended in RPMI1640 w/o phenol red, and added to neutrophils at 2.5 × 105 cells/well for multiplicity of infection (MOI) 5 or 5 × 105 cells/well for MOI 10. Neutrophils were stimulated with 100 nM phorbol 12-myristate 13-acetate (PMA, Sigma Aldrich) for activation and as positive control. Controls of unstimulated neutrophils and each Candida strain without neutrophils were included in each experiment. Luminescence of the Candida only control was subtracted from the luminescence of the coincubation. For the detection of total ROS, 50 µl of RPMI1640 containing 200 µM luminol (Sigma Aldrich) and 16 U HRP (Sigma Aldrich) were added to each well, and chemiluminescence was measured every 2.5 min for 2.5 h in a Tecan Infinite M200 microplate reader. Analyses and determination of the area under the curve were performed with GraphPad Prism 8.4.3.
Fungal survival assay
Freshly isolated neutrophils (see above) were seeded at a concentration of 5 × 104 cells/well in a 96 well plate and incubated for about 1 h at 37°C 5% CO2, to let them attach. Candida cells, opsonized with human serum for 30 min at 37°C, were washed and resuspended in RPMI1640 w/o phenol red, and added to neutrophils at 2.5 × 104 cells/well for MOI 0.5 and co-incubated for 1 h or 3 h, respectively. To determine fungal survival, neutrophils were then lysed with 0.02% Triton-X-100 for 5 min. The lysate containing the fungal cells was rigorously collected from the wells, diluted in PBS, and plated on YPD. After 1–2 days of incubation at 30°C, colony forming units (CFUs) were determined. A control of the infectious dose and Candida cells without the presence of neutrophils was included for each experiment.
Statistical analysis
All experiments were conducted in technical duplicates or triplicates (from which the mean value was calculated) on at least three independent occasions (biological replicates). Different biological replicates included blood or neutrophils from different healthy donors. Diagrams show the mean of the biological replicates with standard deviation (SD). Statistical analyses were done with GraphPad Prism 8.
Results
Stress resistance and environmental survival of Candida auris
In contrast to other pathogenic Candida species, C. auris is often found in the (healthcare) environment, rather than solely associated with the human host [Citation23,Citation24,Citation27,Citation66]. We propose that adaptation of C. auris to environmental stresses allows its survival in a nosocomial setting and may also prime the fungus for survival in certain host niches.
Using drop tests, we compared the resistance of C. auris and the clinically most relevant Candida species (C. albicans, C. glabrata, C. tropicalis, and C. parapsilosis) against oxidative, thermal, and osmotic stress (). Among the compared Candida species, C. auris showed the highest resistance against various stresses, especially growth at 42°C or presence of 10% NaCl. Its growth in the presence of 10 mM H2O2 was comparable to C. glabrata, which is known to have a high tolerance toward oxidative stress ( [Citation67]). Another potential stressor in both environmental and host niches is nutrient limitation. To analyze fungal survival under long-term starvation, we incubated C. auris, the other four common pathogenic Candida species, and the closely related species C. haemulonii in distilled water for 24 days. We observed a remarkable ability of C. auris to withstand these conditions: About 88% of the originally inoculated fungal population was still able to grow and form colonies after 24 days (). This was the case for only about 3% and 21% of the original population of C. haemulonii and C. glabrata cells, respectively. We further analyzed the ability of the different Candida species to grow after drying on a plastic surface without nutrient supply (). C. auris was able to recover and grow following 12 days under such conditions. Only C. parapsilosis showed a comparable ability to resist desiccation, while C. albicans, C. glabrata and C. tropicalis showed no growth after only 3 days. C. haemulonii was severely affected by these conditions ().
Figure 1. C. auris is highly resistant to various stressors in vitro. Drop tests with serial dilutions of C. auris and the clinically most relevant Candida species were performed under various stress conditions to analyze fungal (a) tolerance of oxidative stress, (b) resistance against UV light and high temperature, and (c) resistance against osmotic stress. The highest concentration (left) was 1 × 108 fungal cells/ml. Pictures were taken after 1–2 days of incubation at the indicated temperature and representative examples from at least two biological replicates are shown.
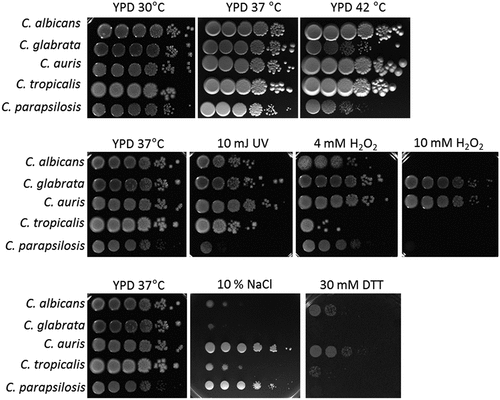
Figure 2. C. auris survives long periods under nutrient-limited, environmental conditions. (a) Survival of Candida spp. in water at 30°C over a time period of 24 days was analyzed by CFU quantification at indicated time points. Results are shown relative to day 0 as 100%. n ≥ 3, mean ± SD; ** p ≤ 0.005, *** p ≤ 0.001 vs. C. auris (b) Candida survival in water over 24 days from (a) is expressed as Area Under Curve (AUC), clearly indicating differences between Candida species. n ≥ 3 (c) Survival of Candida spp. after a dry period of 0, 3, 6 or 12 days was assessed by analyzing re-growth in YPD at 30°C by optical density (OD) measurement at 600 nm over 48 h. n ≥ 3 shown as mean.
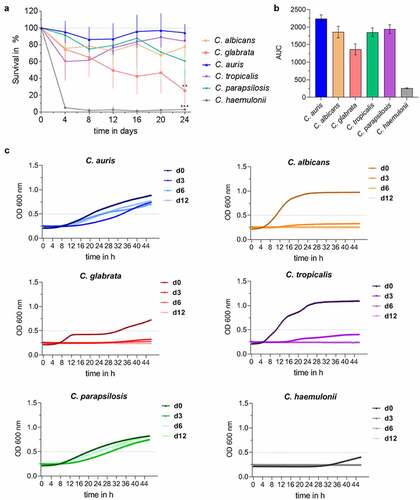
In summary, C. auris showed an extremely high level of resistance to a variety of stresses, likely contributing to survival in the nosocomial environmental and therefore to hospital transmissions and the onset of human infections.
Candida auris is efficiently killed in human blood, but small subpopulations survive
During systemic Candida infections, fungal cells disseminate via the bloodstream throughout the human body. To mimic this critical step of disseminated infections, we applied a previously established ex vivo whole-blood infection model [Citation53,Citation54,Citation68]. We found that C. auris is efficiently killed in human blood of healthy donors (, Figure S1A). Within 60 min post infection (mpi), more than 50% of the C. auris cells were killed, which is comparable to the killing rates of C. albicans and C. glabrata. Despite this efficient clearing, however, after 240 mpi a subpopulation of about 9% of C. auris cells was found to be still alive (C. albicans 12%, C. glabrata 6%). Histological analysis of blood smears during the infection showed no aggregation or replication of C. auris cells and, in contrast to C. albicans, no morphological switch to hyphal growth was detected ( [Citation53]). Furthermore, we observed co-localization of C. auris cells with immune cells. To better characterize which immune cells contribute to killing of C. auris cells in the blood, FITC-labeled C. auris yeasts and immunofluorescence-stained immune cells were analyzed in a flow cytometry-based assay during their interaction in the ex vivo infection model. This analysis showed a rapid association of C. auris with blood leukocytes, in particular neutrophils (). This is in accordance with observations for other Candida spp. like C. albicans and C. glabrata ( [Citation53]). After 240 mpi, only 3% of C. auris cells were not associated with any immune cell (C. albicans ≈ 7%, C. glabrata ≈ 5%). An association with lymphocytes, mainly NK cells, was seen for C. auris at the later time point (0.5% 60 mpi; 4.5% 240 mpi; Figure S1B), comparable to lymphocyte associations of C. albicans and C. glabrata.
Figure 3. C. auris interacts with immune cells and is efficiently killed during ex vivo whole blood infection. (a) Survival of C. auris, C. albicans, and C. glabrata in human whole blood was determined 30, 60, 120, and 240 minutes post infection (mpi) by CFU quantification relative to time point 0 mpi as 100%. n = 6, mean ± SD (b) Exemplary microscopic pictures of blood smears prepared at indicated time points from C. auris infected whole blood. Arrows indicate fungal and human immune cells. 100 x magnification (c) Association of fungal cells with immune cells was determined by fluorescence activated cell sorting (FACS) analyses at 60 mpi and 240 mpi. n = 3, mean ± SD for independent experiments using different donors.
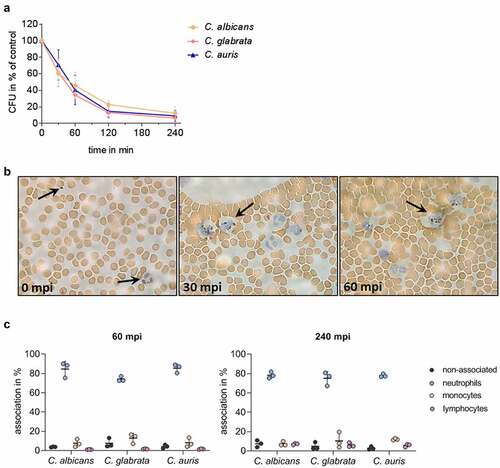
Altogether, these observations demonstrate that C. auris is efficiently killed in human blood and is mostly associated with neutrophils. However, a fraction of the population remained without contact with any immune cells and/or survived in the ex vivo whole-blood infection model.
Dual-species transcriptional profiling of Candida auris-infected human blood
To gain more insights into the mechanisms of fungal adaptation to the blood environment, and counteracting human immune response, dual-species transcriptional profiling by RNA sequencing was performed during C. auris-whole blood infection. We generated C. auris data and used additional-published data sets obtained by the same Standard Operation Procedures for C. albicans, C. glabrata, C. tropicalis, and C. parapsilosis [Citation53], using MRN values and abs(log2FC) ≥ 1.5 (FC > 2.8), adjusted p-value ≤ 0.01 as cutoffs for differential expression of genes.
The human transcriptional response to C. auris infection resembles the response to other Candida species
The transcriptional pattern of human blood cells after infection with each of the five different Candida spp. was mainly governed by the time point post infection, while the infecting species had no measurable influence, as seen by Principal Component Analyses (PCA) ( [Citation53]). After infections with C. auris, similar to the other Candida spp., the number of regulated human genes was initially very low (, Figure S2A). Transcriptional responses of blood cells were predominantly visible from 60 min of co-incubation on, with the number of differentially expressed genes (DEGs) increasing to more than 1,000 DEGs (). Of about 1,300 DEGs seen at 120 mpi and 240 mpi, 793 genes were differentially regulated exclusively at these two time points (), indicating a continuous human response at these later stages of C. auris infection.
Figure 4. The human transcriptional response to C. auris in whole blood infection resembles the response to other Candida species. (a) Principal Component analysis (PCA plot) of the human transcriptome based on MRN values. Different time points (color) and Candida species (icon) are indicated to show similarity (b) Transcriptional kinetics shown by the number of human up- and down-regulated genes at different time points during C. auris infection (compared to 0 min) (c) Venn diagram for Homo sapiens comparing differentially expressed genes (DEGs) of each time point (compared to 0 min) during C. auris infection. (d) Venn diagram for infection with each indicated Candida species comparing human DEGs from infection with the indicated Candida species, considering genes differentially expressed during at least one time point.
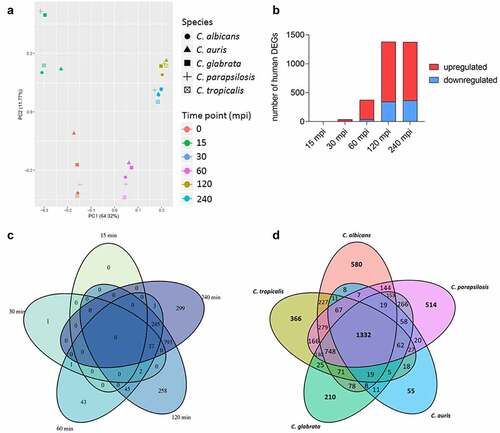
When comparing transcriptional changes induced by C. auris with a published human core response toward infections with C. albicans, C. glabrata, C. tropicalis, and C. parapsilosis [Citation53] we identified a common and conserved human core response of 1,332 DEGs in total to these four Candida spp. and C. auris (, Table S2). In contrast, the number of DEGs specifically regulated in response to any Candida species was much lower, with a maximum of 580 DEGs specifically in response to C. albicans, and only 55 DEGs specific for the response to C. auris (, Table S2).
The common human response seen for C. auris and the other Candida species comprises different infection-relevant aspects, including immune recognition, signaling, and immune effector functions (, Table S3), which is also seen in a gene set enrichment analysis (GSEA) by consistently upregulated inflammatory response (Biocarta LAIR pathway), cytokines (Biocarta CYTOKINE pathway), and IL1R signaling (Biocarta IL1R pathway) sets, among similar others (Figure S2C). Amongst the known fungal-sensing pattern recognition receptors (PRRs [Citation69]), the Toll-like receptor 2 gene (TLR2) was significantly up-regulated during C. auris infection (), while genes coding for TLR5, TLR6, or TLR8 were rather down-regulated. The genes encoding the C-type lectin receptors (CLR) CLEC5a and MRC1 were significantly up-regulated. Furthermore, for the complement receptor CR3 about 10-fold increased transcript levels were found. In line with the activation of the complement system, the complement activating PRR gene, PTX3, as well as the genes encoding the complement component C3 and the complement component receptors C3AR1 and C5AR1 were up-regulated ().
Figure 5. The transcriptional response of blood cells to C. auris is dominated by the expression of immune mediators. (a) Expression of human genes encoding immune cell receptors, cytokines or chemokines, or associated with immune cell signaling or effector functions. Gene expression is shown color-coded as log2FC vs. 0 min and for each time point (15, 30, 60, 120, 240 mpi). More details are given in Table S3. (b) Plasma levels of the pro-inflammatory cytokines IL-1β, IL-6, TNF-α and the chemokine IL-8 240 mpi with C. auris alive or heat-killed (HK) or in an uninfected control sample. n ≥ 5 different donors shown as mean ± SD (c) Expression of selected human cytokines over the time course of C. auris infection. Gene expression is shown as log2FC vs. 0 min, indicated by color intensity.
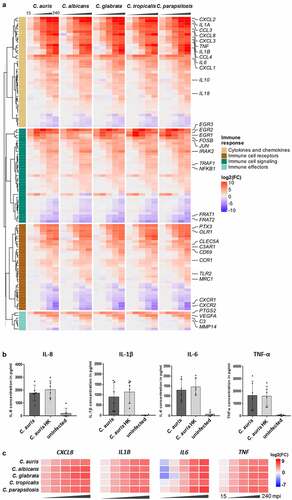
Immune recognition via PRRs leads to the activation of immunological pathways mediating the production and release of immune effectors and to immune cell activities that counteract invading microbes. Up-regulated genes coding for components known to induce the expression of pro-inflammatory cytokines were EGR transcription factor genes (EGR1-3) and genes of the NF-κB pathway, MAPK cascades including JNK, FOS, and AP-1, as well as genes associated with JAK-STAT-signaling (, Figure S2C [Citation70]). The up-regulation of NLRP3 during C. auris infection further indicates the formation of the NLRP3 inflammasome that leads to the secretion of the pro-inflammatory cytokines IL-1β and IL-18 (IL1B > 55-fold up at 60 mpi and > 300-fold up at 240 mpi, IL18 > 4-fold up at 120 mpi; , Table S3 [Citation71]). Furthermore, activation of pro-inflammatory immune responses to C. auris infections, especially cytokine and chemokine production was indicated by the most highly expressed cytokine gene IL1A (> 4,000-fold up at 120–240 mpi), as well as strongly increased expression of IL6 (> 1,000-fold up), TNF (> 170-fold up), and CXCL8 (encoding IL-8, > 180-fold up, ), which was also reflected in the gene set enrichment analysis (Figure S2C). The transcriptional up-regulation of these cytokine genes was further confirmed on the protein level for IL-1β, IL-6, TNF-α, and IL-8 at 240 mpi in C. auris-infected human blood (, Figure S1C). The induction of IL-1β, IL-6, TNF-α, and IL-8 was also seen on transcriptional and protein levels as part of the common human core response against other Candida spp. ( [Citation53]). Pro-inflammatory pathways further lead to the recruitment of other immune cells to the site of infection via chemokines [Citation72]. In line with this, genes encoding chemokines (e.g. CCL3, CCL4, CXCL1, CXCL2 and CXCL3) and their receptors (CCR1, CCR5, CXCR2) were up-regulated in response to C. auris ([Citation72,Citation73]; , Table S3).
Genes encoding the anti-inflammatory cytokines IL-10 [Citation70] and IL1-RA, as well as the TNF-receptor-associated-factor 1 (TRAF1 [Citation74]), that down-regulates the antifungal immune response, were up-regulated only at late time points of C. auris blood infection (). This might indicate a shift toward a down-regulation of inflammatory responses when most of the fungal cells have been killed (). Furthermore, genes encoding matrix metalloproteinases (MMPs) were up-regulated at 240 mpi for all Candida species (mmp14); specific MMP genes like mmp9 were up-regulated during C. auris and C. parapsilosis, but not C. albicans infections (). MMPs are also known to limit inflammation-induced signals by cytokine inactivation [Citation75,Citation76].
The circulating blood cells orchestrate the immune response by communicating not only with each other but also with endothelial cells. Indeed, we found that genes known to activate endothelial cells were up-regulated in response to C. auris (), including PTGS2 (also called COX2, encoding the cyclooxygenase producing prostaglandins), a well-described mediator of pro-inflammatory responses [Citation70,Citation77,Citation78]. Furthermore, genes encoding factors that increase vascular permeability and immune cell migration, like TNF-α and IL-1β or the vascular endothelial growth factor (VEGF) were significantly up-regulated in response to C. auris ( [Citation79,Citation80]). The up-regulation of these genes relevant for the modulation of endothelial cells was comparable for C. auris and the other four Candida species ().
In summary, C. auris is efficiently recognized by the human immune system during blood infection. Association with neutrophils and monocytes was demonstrated on the cellular level. The transcriptional response by these immune cells showed immune recognition by PRRs. This results in pro-inflammatory activities to counteract the fungal infection, with a large overlap in the gene expression patterns of the host in reaction to infections with C. auris, C. albicans, C. glabrata, C. tropicalis, or C. parapsilosis. Together with the observed killing of C. auris in human blood our data suggest an efficient and controlled immune response against this emerging fungal pathogen. However, a substantial portion of C. auris cells can survive 60 min of exposure to human blood and a small fraction was still alive after 240 mpi, indicating fungal adaptation and survival measures.
A pattern of its own: Candida auris species-specific gene expression during whole blood infection
Next, we analyzed the transcriptional adaptation of C. auris (genome reference strain C. auris B8441, representing the South Asian clade I [Citation11,Citation12]) to human blood. It has been previously shown that C. albicans, C. tropicalis, and C. parapsilosis differentially regulate a significant portion of their genome as early as 15 mpi ( [Citation53]). For C. auris, about 15% of the transcriptome was differentially regulated at early time points, which is more than for C. glabrata (1.3–8.2% over the time course, [Citation53]). The numbers of genes that were significantly up- and down-regulated in C. auris were comparable (, Figure S2B [Citation53]). The genomes of C. auris and the other four Candida species share ≈ 3,700 orthologues amongst which only 87 were differentially regulated in all species at any time point during infection (). This small set of fungal core response genes includes heat shock protein genes like HSP104, HSP78, and SBA1 which were all up-regulated. They thereby indicate stress adaptation to the hostile blood niche via heat shock proteins in all five Candida species (, Table S5). However, more genes were species-specifically than commonly regulated (; except for C. glabrata). Investigating the time course of C. auris DEGs, only 42 DEGs were found to be regulated at all time points (). Many of these genes are associated with transmembrane transport, for example PHO84, HGT10 and OPT2 (, Figure S3). The C. auris genome contains a substantial number of transporter genes [Citation32,Citation81]. This includes the gene encoding the multidrug resistance efflux pump Mdr1, which is known to play a key role for resistance to azoles in C. albicans and also C. auris [Citation32,Citation82]. During whole blood infection MDR1 expression was highly up-regulated at all time points (>12 – 34-fold up), similar to other MDR family members (e.g. B9J08_001319 > 3 – 5-fold up; , Table S6). Notably, C. albicans, C. tropicalis, and C. parapsilosis did not up-regulate their orthologous transporter genes to the same extent, and these were also not found to be up-regulated over the whole course of infection.
Figure 6. Species-specific gene expression of C. auris during whole blood infection. (a) The transcriptional changes during whole blood infection are shown as % of the genome for each Candida species at the indicated time points. The total number of predicted open reading frames (ORF) is given in brackets for each species (b) Transcriptional kinetics shown by the number of up- and down-regulated C. auris genes at different time points during whole blood infection (compared to 0 min). (c) Venn diagram for C. auris comparing fungal differentially expressed genes (DEGs) of each time point (compared to 0 min) during whole blood infection. (d) Venn diagram comparing fungal DEGs for each indicated Candida species over the time course of whole blood infection, considering gene orthology and differential expression for at least one time point.
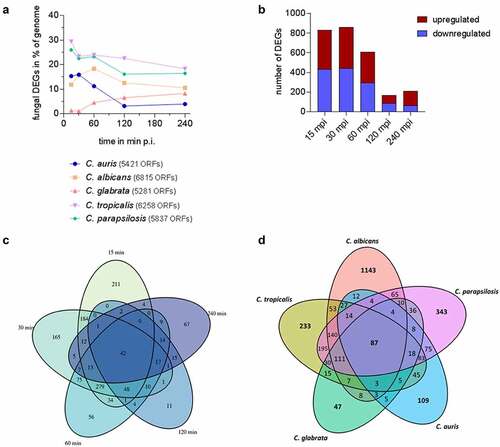
Figure 7. A pattern of its own: C. auris specific transcriptional alterations in response to human blood. Expression of selected fungal genes over the course of infection (15, 30, 60, 120, 240 mpi) with the indicated Candida species, associated with (a) protein folding, translation, the cell wall, oxidative stress or related to transport across the membrane, metal homeostasis, adhesion, or (b) metabolism. Gene expression is shown for orthologous genes color-coded as log2FC vs. 0 min for each time point. Genes without orthologues (or best hit according to BLASTP) are shown in gray. More details are given in Table S6.
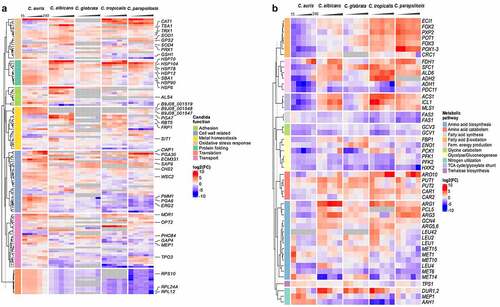
Furthermore, genes coding for amino acid (GAP4) or ammonium transporters (MEP1) were up-regulated in C. auris (>2 or 8-fold up at 60 mpi, respectively; ). These transporters may be required for the uptake of essential nutrients and metabolic adaptations. However, the mode of metabolic adaptation seems to differ between Candida species. Accordingly, we observed no up-regulation of the glyoxylate cycle genes, ICL1 and MLS1, or genes associated with fermentative energy production (up-regulation of ADH2 and ALD6) in C. auris, in contrast to C. albicans (). Furthermore, genes involved in β-oxidation (CRC1, POX1-3) were down-regulated in C. auris over the entire course of infection, in contrast especially to C. parapsilosis (up-regulation of ECI1, FOX2, FOX3, POT1, and PXP2 – all involved in β-oxidation), indicating differences in the utilization of fatty acids as an energy source (, Table S6).
While only a small number of C. auris genes were differentially expressed at all time points during blood infection, several genes were expressed at more than one time point and the C. auris transcriptome can be separated into characteristic early and late responses, with the early 15, 30, and 60 mpi time points having nearly 280 genes in common (, Table S2). At these early time points, most of the up-regulated genes were involved in translation, like RPL12, RPL24A, and RPS10, and associated processes such as chaperone-mediated protein folding (Figure S3). While the up-regulation of genes involved in translation was unique for C. auris among the tested Candida species (), transcriptional regulation of protein folding events was commonly found. However, the regulated genes used in these processes were diverse (based on the orthologue data). For example, HSP6 was solely up-regulated in C. auris (> 8.5-fold), while up-regulation of HSP12 was seen in C. albicans and C. parapsilosis, but not in C. auris (). In addition, up-regulation of HSP90 seems to be relevant for C. auris during all stages of blood infection (> 14-fold at early time points; ), while there was no differential regulation of HSP90 in C. albicans or C. glabrata. C. albicans, C. tropicalis, and C. parapsilosis, on the other hand, all up-regulated HSP70 encoding an important Hsp, at least for C. albicans virulence [Citation83].
At later time points (120 and 240 mpi), all Candida species showed up-regulation of genes associated with cell wall organization and biosynthesis, however, the different species used different sets of genes in this category (, Figure S3). For example, CWP1 (encoding Cell Wall Protein 1) is increasingly expressed over the course of infection in C. auris (> 470-fold at 240 mpi) and C. glabrata (> 13-fold at 240 mpi), while there is no orthologue in C. albicans. Similarly, the gene coding for the fungal cell wall protein Pga30 was highly up-regulated in C. auris (> 119-fold) and C. parapsilosis (> 70-fold), but not in C. albicans (, Table S6). Among the more than 100 DEGs only up-regulated in C. auris (, Table S4), a large portion comprised cell membrane- and cell wall-associated genes. Namely, genes related to ergosterol biosynthesis (ERG2, ERG6) or chitin biosynthesis (CHS3, CHS8) and other related genes (PMM1, FEN1, ECM17, WSC2) were specifically up-regulated in C. auris ().
Species-specific differences also became obvious in terms of iron acquisition during whole-blood infection, as orthologous genes for ferric reductases (FRP1, FRP2), hemoglobin utilization (RBT5, PGA7), and siderophore transport (SIT1) were differentially regulated in C. auris and C. albicans during infection ().
While C. auris does not have orthologues of key C. albicans virulence genes, like ECE1 or PRA1 [Citation12], C. auris expresses genes without orthologues in any of the other four analyzed Candida species. Of the 659 C. auris-specific genes analyzed in this study, over half of them (368 out of 659) were differentially expressed during at least one time point of infection (Table S7). Of note, 96 of the C. auris-specific up-regulated DEGs at 240 mpi were of unknown function, and others were associated with transmembrane transport (Figure S3). This includes genes potentially relevant for iron homeostasis and acquisition during blood infection, for example, B9J08_001548, B9J08_001547, and B9J08_001519. These genes have no clear orthologues but were found to be similar (by NCBI BLASTP analyses [Citation84],) to genes encoding the siderophore transporter Sit1 (), and genes B9J08_000568 and B9J08_001951 were found to be similar to genes encoding the ferric reductase Fre3 and Csa1, a member of a hemoglobin-receptor family of C. albicans.
In summary, C. auris showed numerous transcriptional adaptations, including changes in membrane transport and the cell wall, likely enabling the fungus to cope with host-derived stresses and survive in the blood niche. Of note, this includes the up-regulation of known drug-resistance genes, which may contribute to drug resistance in a clinical setting.
Candida auris oxidative stress response during whole blood infection
Blood-borne immune cells have to kill invading microbes and prevent microbial dissemination. One killing strategy of immune effector cells like neutrophils is the production of reactive oxygen species (ROS). This oxidative burst can be primed by cytokines such as TNF-α or chemokines such as IL-8. Increased expression of the corresponding genes (cxcl8, tnf) was indeed detected during the C. auris-blood infection (). The activation of neutrophils was also indicated by the up-regulation of the early activation marker gene CD69 (> 5-fold up from 30 mpi on), and the gene encoding CD83 (> 12-fold up from 30 mpi on) both also known to be activated in neutrophils during C. albicans infections [Citation39].
Contact to neutrophils, the most abundant immune cells in blood, causes fungal oxidative stress responses in C. albicans [Citation68,Citation85] and C. glabrata [Citation86,Citation87]. Consequently, we compared the expression of C. auris oxidative stress response genes during blood infection with the expression of correlated genes for other Candida species ().
The key genes encoding detoxifying enzymes of C. albicans are the superoxide dismutase genes SOD4 and SOD5, which are all up-regulated during blood infection [Citation53,Citation68]. In contrast, the C. auris superoxide dismutase genes (SOD1 and SOD4; no orthologue for SOD5) were only weakly up-regulated (). While the catalase gene CAT1 and its orthologues were up-regulated in C. albicans, C. glabrata, C. tropicalis and C. parapsilosis, this was not observed in C. auris (). Furthermore, the glutathione system (including GSH1, GPS2, GST1, GPXs, and GTTs [Citation85,Citation88,Citation89]) contributes to ROS detoxification in C. albicans and members of this gene set are up-regulated in C. albicans, C. tropicalis, and C. parapsilosis. In contrast, the corresponding orthologous genes (as far as they exist) are not up-regulated in C. auris or C. glabrata (), suggesting a minor role (at most) of the glutathione system in the oxidative stress response of these two species.
However, we observed that PRX1 encoding the putative thioredoxin peroxidase was significantly up-regulated at early time points in C. auris (> 30-fold up) but not in C. albicans and C. parapsilosis, putatively contributing to ROS detoxification. A similar species-specific expression was visible for other components of the thioredoxin system (TSA1, TRX1, and TRR1) (). Furthermore, the C. auris orthologues (B9J08_003622 and B9J08_000834) of C. albicans orf19.7085, which has been described to be oxidative stress-induced [Citation90], were highly up-regulated during infection as were the orthologous genes in C. tropicalis and C. parapsilosis ().
Taken together, genes for ROS detoxifying enzymes and systems seem to be present in C. auris and other tested Candida species. However, their expression patterns during blood infections were species-specific, suggesting an individual strategy for C. auris to cope with immune cell-derived oxidative stress.
Candida auris interacts with human neutrophils
Since neutrophils and their oxidative burst are key players during Candida bloodstream infections ( [Citation53,Citation68]), we investigated the direct interaction of C. auris with neutrophils in comparison to the other Candida species. Neutrophils clearly showed ROS production in response to C. auris and all tested Candida species (). However, as suggested by the transcriptional profiles, all the investigated Candida species were able to detoxify ROS produced by PMA-stimulated neutrophils ( [Citation91]). We then analyzed the killing of C. auris by neutrophils. Only 15% of C. auris cells survived after 60 mpi and the overall viability of fungal cells were further decreased to 2% 180 mpi, underlining the key role of neutrophils in the blood against C. auris infections ().
Figure 8. Human neutrophils recognize C. auris in vitro and respond with oxidative burst. (a) Human neutrophils were infected with different Candida strains at an MOI of 5. Phorbol 12-myristate 13-acetate (PMA) was used as a positive control. Results are shown as Area Under Curve (AUC) based on the ROS measurement presented in (b) n = 8, mean ± SD (B) ROS measurement over the course of co-incubation of different Candida strains with human neutrophils in vitro. MOI 5; n = 8; *** p ≤ 0.001 vs. C. auris (c) Human neutrophils were treated with PMA and in parallel infected with the indicated Candida spp. at an MOI of 10 to detect fungal ROS detoxification. Results are shown as AUC of the ROS measurement. n ≥ 7; *** p ≤ 0.001 vs. PMA.
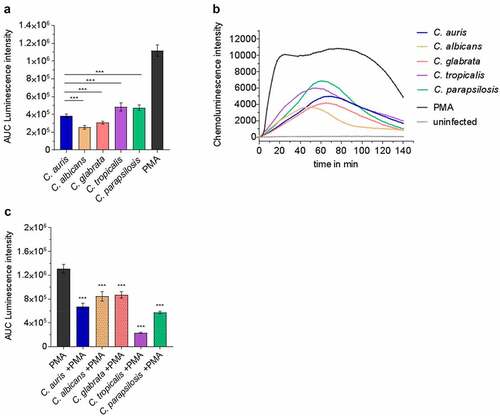
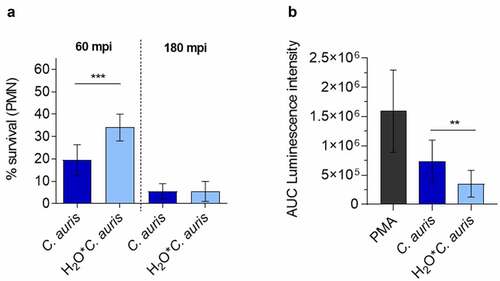
Figure 9. C. auris survival of neutrophil infection. (a) Survival of C. auris after 60 min or 180 min co-incubation with human neutrophils in vitro, determined by CFU quantification. n = 6, mean ± SD; *** p≤ 0.001 (b) Induction of ROS in human neutrophils after co-incubation with C. auris compared to C. auris pre-starved in water (H2O*C. auris) over 6 days. n = 6, mean ± SD; ** p≤ 0.005 .
Effects of the environmental preconditions on the survival rate of C.auris
Although we did not observe greater resistance of C.auris to killing in whole blood or by human neutrophils compared to other clinically relevant Candida species, it is the Candida species of major concern for hospital outbreaks. We next questioned whether the high-stress resistance and the high capacity of C.auris to survive harsh environmental conditions and nutrient limitation () may be relevant for the transfer from the environment to a patient. To analyze the influence of previous environmental conditioning on interactions with blood cells, we starved C. auris in water and subsequently determined fungal survival after co-incubation with human blood-derived neutrophils in vitro. We observed a significantly increased survival rate at 60 mpi after pre-starvation in water (). In contrast, pre-starvation did not affect survival rates at the later stages of infections. Potentially, growth under harsh environmental conditions changes fungal cell properties, which may dampen antifungal activity by host cells. In fact, ROS production by neutrophils was significantly less pronounced after infection with pre-starved C. auris cells compared to standard conditions ().
These data suggest that the environmental conditions C.auris faces prior to infection can modulate the initial interaction between human neutrophils and early bloodstream fungal survival.
Discussion
Infections with Candida species are considered a global public health threat, especially in the nosocomial setting [Citation92,Citation93]. While C. albicans is still the most common cause of life-threatening candidiasis and the best-studied Candida species, the incidence of and interest in non-albicans species such as C. tropicalis, C. parapsilosis, C. glabrata, and C. auris is increasing [Citation94]. Lately, C. auris infections became of particular public concern due to outbreaks in health care settings in different countries, the unusual high intrinsic antifungal resistance of this fungus, and by its spread in specialized COVID-19 ICUs during the current pandemic [Citation95,Citation96]. A better understanding of the pathogenicity of C. auris is therefore vital.
In our study, we shed light on the interplay between C. auris and human blood cells during ex vivo blood infection by characterizing cellular interactions with blood cells, the immune response and the transcriptome of both host and fungal cells using dual-species RNA-Seq. Our study reveals a human response to C. auris infection that is dominated by early immune system activation, and which is comparable to infections with other Candida species. On the fungal side, the transcriptional changes of C. auris during whole-blood infection were clearly different in comparison to other medically important Candida species, indicating a specific response of C. auris to blood exposure that does not resemble known Candida species.
While C. auris does not appear to be more pathogenic than other Candida species in our model, it has caused hospital outbreaks with severe nosocomial transmission [Citation24,Citation31,Citation95,Citation97,Citation98]. Our analyses of C. auris’ resistance against stress and harsh in vitro conditions showed a remarkable ability of this fungus to survive in the environment for a long time. This provides a possible explanation for the scale of nosocomial infections, as C. auris may survive better on many different hospital surfaces than other Candida species. Importantly, the same characteristics may not only facilitate nosocomial survival and persistence, but also resistance to host-derived stressors during the early stages of infection.
Human responses to C. auris blood infection
During blood infection, the vast majority of infecting C. auris cells were associated with neutrophils, similar to C. albicans and C. glabrata, underlining the importance of this interaction in fungal blood infections in general ( [Citation53,Citation99]). Furthermore, some fungal cells associated with monocytes ( [Citation42,Citation53,Citation100]).
The human transcriptional response is likely derived predominantly from these blood-resident immune cells. These immune cells (neutrophils, monocytes) have a preformed repertoire of immune mediators, explaining an initially limited transcriptional response to infections with any of the five Candida species, clearly increasing after 60 mpi (, Figure S2A [Citation53]). Our transcriptional data indicate a recognition of C. auris by these cell types via PRRs, including the C-type lectin receptors (CLRs) MRC1 and CLEC5a, the complement receptor CR3, and the Toll-like receptor TLR2 (). This activates MAPK-related signaling, NF-κB-signaling, and EGR regulators that lead to pro-inflammatory activities, including the expression and secretion of cytokines like IL-1α, IL-1β, IL-6, IL-13, and TNF-α, as well as chemokines and other anti-fungal responses (). Similarly, CLR-associated TNF-α, IL-6, IL-1β, and chemokine expression was reported by isolated PBMCs in response to C. auris [Citation37]. In our blood model, comparison of C. auris infections with previous data on Candida species demonstrated that the transcriptional response on the host side was largely conserved and mainly dependent on the time point of infection rather than on the infecting species – in line with our previous four-species observations ( [Citation53,Citation54,Citation68]). The common human response even extends to infections with C. albicans cells, although these transition to hyphae during blood infection [Citation53,Citation54], while the other four species, including C. auris, remain in the yeast morphology ( [Citation53]). Niemiec et al. described a morphology-independent neutrophil response toward C. albicans [Citation39], which resembled the response to infections with C. auris in our study, including the up-regulation of PTX3, NLRP3, EGR1-3, TNF, and chemokine genes (). An efficient early neutrophil activation by non-filamentous C. albicans strains (when components of the complement system are present) has been reported earlier [Citation101], which is also the case in our blood model.
On the host side, we therefore show that the antifungal defense is effective in fungal killing, dominated by pro-inflammatory responses, and largely conserved against several Candida species.
C. auris responses to human blood
In line with the human immune response toward Candida cells, the majority of C. auris cells are cleared in our blood infection model over 240 min (). However, a small fungal subpopulation survived, and in the in vivo situation an escape from the bloodstream would be expected within this timeframe [Citation102,Citation103]. Therefore, our data provide experimental evidence that a portion of C. auris cells can survive long enough to escape from the bloodstream as a prerequisite to colonize organs. Our data indicate that C. auris is in fact able to proliferate in murine kidney and liver homogenates, as well as in organ-simulating medium, comparable to, or at even higher rates than other Candida spp. (Figure S4).
Transcriptional profiling was used to elucidate the contribution of C. auris genes and potential virulence factors to adaptation and survival in blood. However, gene functions have so far been mainly ascribed to orthologue information, rather than functional characterization in C. auris itself. Nevertheless, the genome of the C. auris strain B8441 has been sequenced and annotated by comparative genomic analyses, which showed conservation of, amongst others, genes associated with virulence within the Candida clade [Citation12,Citation32,Citation94].
At the transcriptional level, metabolic adaptations to the bloodstream environment were observed for C. auris as well as for other Candida species. However, the regulation of genes in catabolic and anabolic pathways differed (). For example, C. tropicalis and C. parapsilosis up-regulated β-oxidation, and C. albicans, C. tropicalis, and C. parapsilosis up-regulated glyoxylate cycle genes, which was not the case for the orthologous C. auris genes throughout the course of infection (). Contact to neutrophils was also previously described to induce amino acid synthesis, e.g. the methionine biosynthesis pathway in C. albicans [Citation85,Citation104]. We found genes of this pathway (MET1, MET10) up-regulated in C. albicans and C. auris at early stages of blood infection, while this was not seen in C. parapsilosis ( [Citation53]). As amino acids are present in blood, they might be acquired via transporters to serve as carbon and nitrogen sources [Citation105]. Especially in the C. auris genome the number of transporter genes, like the oligopeptide transporter family (OPT genes) that enable the uptake of small peptides, is expanded [Citation12]. The up-regulation of several C. auris amino acid transporter genes (GAP4, OPT1, B9J08_004537 (OPT2), PUT1; ), suggests that amino acids are indeed imported as a nutrient source during blood infection. Overall, these data indicate the use of different carbon sources by the different Candida species during blood infection. This may be explained by variable nutrient acquisition and metabolic strategies evolved in the individual host- or environment-associated natural reservoirs of each fungus [Citation105].
Further interspecies differences are expected due to the fact that distinct virulence traits like the yeast-to-hypha transition and the peptide toxin candidalysin of C. albicans are not present in C. auris [Citation12]. Filamentation contributes to C. albicans survival in blood [Citation53] and might account for the moderately higher survival of C. albicans compared to C. auris (). Other genes encoding virulence-associated factors of C. albicans, like secreted proteases (Saps) and lipases (Lips) have orthologues in the C. auris genome [Citation12,Citation94]. The LIP10 and the SAP9 orthologues of C. auris as well as two other genes with similarity to SAP9 (B9J08_003911, B9J08_005335) were significantly up-regulated during incubation in blood () and may contribute to the interaction with blood cells. For example, C. albicans Sap9 has been shown to be required for neutrophil activation including chemotaxis, ROS formation, and NETosis [Citation106,Citation107], and its C. auris counterparts may have similar roles.
Furthermore, C. auris harbors an expanded set of transporter genes, including several members of the ABC and MFS gene families [Citation12,Citation81,Citation108]. Interestingly, genes of the expanded MFS class, known for their role in antifungal resistance [Citation12,Citation94], seem to be relevant during C. auris blood infection as MDR1, but also TPO3 and related genes (B9J08_000368, B9J08_003036), were highly up-regulated in our study (). This regulation pattern is unique to C. auris compared to the other four analyzed Candida species, suggesting a specific role of these transporters during C. auris during blood infection. The physiological role of MFS-MDR transporters might include the transport of lipids, ions, or other small metabolites during adaptations to the (host) environment [Citation109]. In C. lusitaniae, Mdr1 has been shown to confer resistance to bacterial and host products like methylglyoxal or hydrogen peroxide [Citation110,Citation111] and MDR1 expression in C. albicans was found under oxidative stress conditions [Citation109,Citation112]. Moreover, the ABC transporter Afr1 of Cryptococcus neoformans has been shown to be relevant for virulence by mediating protective effects against phagocytes [Citation113]. While these studies suggest that transmembrane transporters can contribute to fungal fitness in challenging environments, the up-regulation of members of MFS transporter genes like MDR1 in C. auris during blood infection may at least partially contribute to C. auris azole resistance in patients. This up-regulation of genes involved in counteracting azoles in the blood niche might prepare C. auris for subsequent antifungal therapy, contributing to clinical failures to clear this fungus with this class of drugs during infections.
Another gene family that is expanded in C. auris comprises SIT (siderophore transporter) genes, related to iron uptake and homeostasis [Citation12]. The up-regulation of SIT1 and related genes (B9J08_001519, B9J08_001548, B9J08_001547) during blood infection highlights the relevance of these processes for C. auris in an environment which is virtually free of unbound iron ions (). An association of the previously mentioned expanded C. auris gene families with host interactions and fungal pathogenicity is further in line with earlier observations on gene expansion in C. albicans [Citation114].
Variations in the cell surface structures between different species likely influence immune recognition and response [Citation52]. Unique mannan structures as well as different chitin levels of the C. auris cell wall compared to other Candida species have been described [Citation37,Citation52,Citation115]. During blood infection, expression of genes associated with cell wall biogenesis and remodeling was different for C. auris in comparison to the other Candida species (). Especially genes related to ergosterol and chitin synthesis, as well as PGA genes, were primarily up-regulated in C. auris. This suggests that C. auris modulates its cell wall in response to the host differently than other Candida spp., which in turn might contribute to the few C. auris-specific transcriptional changes in the host response observed in this study.
Taken together, C. auris’ transcriptional adaptations to the blood niche especially relate to metabolism, cell wall remodeling, and acquisition of micronutrients – processes that are also regulated by other Candida species during blood infection. However, much of the transcriptional patterns and kinetics were species-specific and suggest an independent strategy for C. auris to deal with the blood environment. Of note, this includes the up-regulation of genes, especially encoding transporters, known to be associated with azole resistance.
Neutrophils as the key player during C. auris blood infections
The interaction of C. albicans with blood cells has been investigated with isolated immune cells as well as in blood models [Citation40,Citation41,Citation53,Citation68]. Fradin et al. demonstrated that the transcriptional profile of C. albicans in blood is dominated by transcriptional responses to neutrophils [Citation68]. The generation of reactive oxygen species (ROS) is a central defense mechanism of neutrophils against fungal pathogens [Citation40,Citation41,Citation116,Citation117]. The generation of ROS by neutrophils was also seen after contact to C. auris (). It appeared slightly delayed compared to C. albicans (), which is known to be quickly recognized and attacked by neutrophils [Citation42], but seemed comparable to the other Candida spp.
In response, fungal cells express a plethora of genes to counteract oxidative stress [Citation68]. However, the individual oxidative stress adaptations differed (). This can be explained in part by a different genetic repertoire, for example, C. albicans has an evolutionarily expanded set of superoxide dismutase (SOD) genes [Citation85]. C. auris does not have a orthologue of SOD5 but does possess a SOD4 orthologue which was up-regulated during blood infection, suggesting a contribution of at least one Sod to detoxify O2− in C. auris ( [Citation118]). C. auris orthologous genes of other antioxidant mechanisms like catalases (CAT1) or the glutathione/glutaredoxin (GSH, GPX, GST) system [Citation85] were not up-regulated during blood infection, while they are used by C. albicans and C. glabrata under oxidative stress conditions [Citation67]. However, the thiol-specific peroxidase Tsa1 of the thioredoxin system (TSA1, TRX1, TRR1), known to protect C. albicans against ROS [Citation85], also seems to be relevant for C. auris during oxidative stress in blood, as the orthologous gene was significantly up-regulated (). Furthermore, the putative peroxidase gene PRX1 and numerous genes encoding heat shock proteins and enolases, shown to be involved in fungal oxidative stress response [Citation119], showed high mRNA copy numbers in C. auris during blood infection (). Accordingly, the neutrophils’ ROS production can be detoxified by C. auris as well as the other four Candida species ().
In our study, substantial killing of C. auris by isolated human neutrophils was observed 60 mpi and further increased at 180 mpi, in line with the induction of ROS (, ). In contrast, Johnson et al. reported inefficient killing of C. auris by human neutrophils, which is concordant with reduced ROS generation compared to C. albicans [Citation36]. This may partially be explained by the use of different C. auris strains or more likely by the fact that infections in our study include opsonization with human serum, which was shown to be critical for neutrophil activation in our experimental settings (Figure S5). The crucial role of opsonization for efficient host cell response and killing of Candida species by phagocytes – as seen for C. auris-neutrophil interactions in our study – is in agreement with previous studies [Citation120–122], including C. auris-infected PBMCs [Citation37]. Along the same line, ROS production by neutrophils was observed with serum-opsonized, heat-killed C. auris cells (Figure S5 [Citation37]) and Bruno et al. demonstrated myeloperoxidase (MPO) production by neutrophils in C. auris-infected mice, demonstrating in vivo activation of neutrophils during C. auris infection [Citation37].
Combined transcriptional and cellular observations of C. auris during blood infection and interactions with isolated neutrophils demonstrated that C. auris experiences oxidative stress. Despite oxidative stress response and high H2O2-resistance, C. auris is efficiently killed in blood and by neutrophils in vitro, suggesting an effective combination of oxidative burst with other anti-microbial mechanisms in these in vitro and ex vivo settings and thus potentially also in vivo.
C. auris stress resistance impacts environmental survival and virulence
C. auris infections are often reported as outbreaks in health-care settings, which seem to be linked to fungal persistence in the hospital environment [Citation23,Citation95] and intra- or inter-hospital C. auris transmission [Citation9,Citation24,Citation97,Citation123,Citation124]. Nosocomial transmission is well known for C. parapsilosis, while infections with C. albicans, C. glabrata or C. tropicalis are thought to mainly originate from endogenous fungal populations [Citation125–128].
In our study, long-term incubation of Candida cells in water () lacking any nutrients or on plastic surfaces, similar to material found in hospital settings () revealed a remarkable ability of C. auris to survive harsh environmental conditions over several days or even weeks – in contrast to all other tested Candida species, except the other majorly hospital-environmental species C. parapsilosis [Citation129], which is in agreement with previous studies on C. auris and C. parapsilosis [Citation23,Citation130,Citation131]. Thus, we have shown and confirmed that C. auris can survive and persist for long periods outside of the human host, and that nosocomial C. auris infections can therefore originate from environmental sources, such as contaminated medical devices or hands of health care workers, without prior colonization of the infected host [Citation95,Citation126]. This increases the risk of transmission in healthcare settings [Citation24,Citation108,Citation132] and explains why hospital outbreaks are described for C. auris and C. parapsilosis rather than for the other Candida species [Citation16,Citation17,Citation131].
We propose that features, which increase environmental survival of C. auris, including resistance to environmental stressors, like UV light, dryness, or salt, might also have effects on the interaction with the human host and contribute to C. auris virulence. We therefore analyzed the interaction of C. auris cells pre-grown under harsh environmental conditions with human neutrophils (). In fact, these experiments suggest that environmental conditions can significantly influence subsequent infections with immune cells at early stages. Such impact on C. auris-host interactions after environmental starvation or other stressful conditions might be explained by differences in basal gene expression, storage levels of essential micronutrients or cell surface characteristics. The latter would likely influence recognition and responses by the innate immune system, as demonstrated for C. albicans in response to pH or defined nutrient sources [Citation133–135]. Finally, it is possible that such pre-conditioned C. auris cells may be more suitable to colonize the skin [Citation28].
We conclude that the remarkable ability of C. auris to resist environmental stressors and to adapt to harsh long-term conditions can impact environmental survival, the existence of hospital pools of viable and infectious cells and the initial phase of infections.
Supplemental Material
Download Zip (4 MB)Acknowledgments
We thank the Mycotic Disease Branch of the CDC Atlanta (USA) and Neil Gow (University of Exeter, UK) for providing C. auris strains used in this study.
We kindly thank all voluntary blood donors and all who contributed to blood drawing and neutrophils isolation, especially Marina Pekmezovic, Marisa Valentine, Mark S. Gresnigt, and Rita Müller from the MPM department/HKI Jena. We further thank Till Kalkreuter from the Microbial Pathogenicity Mechanisms (MPM) department/HKI Jena for assisting with bioinformatic evaluation of data and Christine Dunker from the Microbial Immunology (MI) group/HKI Jena for the protocol analyzing fungal growth in mouse organ homogenates. Furthermore, thanks go to Annika König and Jakob Sprague for critical reading of the manuscript.
This work was partially funded by the German Research Foundation (Deutsche Forschungsgemeinschaft–DFG) project Hu 532/20-1, project C1, and INF within the Collaborative Research Centre (CRC)/Transregio (TR) 124 FungiNet.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The RNA-Seq data that support the findings of this study are openly available in NCBI’s Gene Expression Omnibus under the GEO record GSE179000and GSE114180. https://www.ncbi.nlm.nih.gov/geo/
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007;45(4):321–346.
- Wisplinghoff H, Ebbers J, Geurtz L, et al. Nosocomial bloodstream infections due to Candida spp. in the USA: species distribution, clinical features and antifungal susceptibilities. Int J Antimicrob Agents. 2014;43(1):78–81.
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20(1):133–163.
- Guinea J. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect. 2014;20(6):5–10.
- Turner SA, Butler G. The Candida pathogenic species complex. Cold Spring Harb Perspect Med. 2014;4(9):a019778.
- Kean R, Brown J, Gulmez D, et al. Candida auris: a decade of understanding of an enigmatic pathogenic yeast. J Fungi (Basel). 2020;6(1). DOI:10.3390/jof6010030.
- Chowdhary A, Sharma C, Meis JF. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017;13(5):e1006290.
- Kullberg BJ, Arendrup MC. Invasive Candidiasis. N Engl J Med. 2015;373(15):1445–1456.
- Sears D, Schwartz BS. Candida auris: an emerging multidrug-resistant pathogen. Int J Infect Dis. 2017;63:95–98.
- Skrzypek MS, Binkley J, Sherlock G. Using the Candida genome database. Methods Mol Biol. 2018;1757:31–47.
- Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous emergence of multidrug-resistant candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64(2):134–140.
- Munoz JF, Gade L, Chow NA, et al. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun. 2018;9(1):5346.
- Forsberg K, Woodworth K, Walters M, et al. Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med Mycol. 2019;57(1):1–12.
- Satoh K, Makimura K, Hasumi Y, et al. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53(1):41–44.
- Muñoz JF, Welsh RM, Shea T, Batra D, Gade L, Howard D, Rowe LA, Meis JF, Litvintseva AP, Cuomo CA , et al. Clade-specific chromosomal rearrangements and loss of subtelomeric adhesins in Candida auris. Genetics. 2021 May 17; 218(1): iyab029. doi:10.1093/genetics/iyab029.
- Chow NA, Gade L, Tsay SV, et al. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis. 2018;18(12):1377–1384.
- Welsh RM, Sexton DJ, Forsberg K, et al. Insights into the unique nature of the East Asian clade of the emerging pathogenic yeast Candida auris. J Clin Microbiol. 2019;57(4). DOI:10.1128/JCM.00007-19.
- Jeffery-Smith A, Taori SK, Schelenz S, et al. Candida auris: a review of the literature. Clin Microbiol Rev. 2018;31(1). DOI:10.1128/CMR.00029-17.
- Casadevall A, Pirofski LA. Accidental virulence, cryptic pathogenesis, martians, lost hosts, and the pathogenicity of environmental microbes. Eukaryot Cell. 2007;6(12):2169–2174.
- Jabra-Rizk MA, Kong EF, Tsui C, et al. Candida albicans pathogenesis: fitting within the host-microbe damage response framework. Infect Immun. 2016;84(10):2724–2739.
- Polvi EJ, Li X, O’Meara TR, et al. Opportunistic yeast pathogens: reservoirs, virulence mechanisms, and therapeutic strategies. Cell Mol Life Sci. 2015;72(12):2261–2287.
- Limon JJ, Skalski JH, Underhill DM. Commensal Fungi in Health and Disease. Cell Host Microbe. 2017;22(2):156–165.
- Welsh RM, Bentz ML, Shams A, et al. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol. 2017;55(10):2996–3005.
- Schelenz S, Hagen F, Rhodes JL, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control. 2016;5(1):35.
- Horton MV, Johnson CJ, Kernien JF, et al. Candida auris forms high-burden biofilms in skin niche conditions and on porcine skin. mSphere. 2020;5(1). DOI:10.1128/mSphere.00910-19.
- Uppuluri P. Candida auris biofilm colonization on skin niche conditions. mSphere. 2020;5(1). DOI:10.1128/mSphere.00972-19
- Arora P, Singh, P, Wang, Y, Yadav, A, Pawar, K, Singh, A, Padmavati, G, Xu, J, Chowdhary, A, et al. Environmental isolation of Candida auris from the coastal wetlands of Andaman Islands, India. mBio. 2021 Mar 16;12(2): e03181–20. doi:10.1128/mBio.03181-20.
- Huang X, Hurabielle C, Drummond RA, et al. Murine model of colonization with fungal pathogen Candida auris to explore skin tropism, host risk factors and therapeutic strategies. Cell Host Microbe. 2021;29(2):210–221 e6.
- Ben-Ami R, Berman J, Novikov A, et al. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis. 2017;23(1). DOI:10.3201/eid2302.161486.
- Heaney H, Laing J, Paterson L, et al. The environmental stress sensitivities of pathogenic Candida species, including Candida auris, and implications for their spread in the hospital setting. Med Mycol. 2020;58(6):744–755.
- Biswal M, Rudramurthy SM, Jain N, et al. Controlling a possible outbreak of Candida auris infection: lessons learnt from multiple interventions. J Hosp Infect. 2017;97(4):363–370.
- Chatterjee S, Alampalli SV, Nageshan RK, et al. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genomics. 2015;16(1):686.
- Kim SH, Iyer, KR, Pardeshi, L, Muñoz, JF, Robbins, N, Cuomo, CA, Wong, KH, Cowen, LE, et al. Genetic analysis of Candida auris implicates HSP90 in morphogenesis and azole tolerance and CDR1 in azole resistance. mBio. 2019 Jan 29;10(1): e02529–18 doi:10.1128/mBio.02529-18.
- Borman AM, Szekely A, Johnson EM. Comparative pathogenicity of United Kingdom isolates of the emerging pathogen candida auris and other key pathogenic Candida species. mSphere. 2016;1(4). DOI:10.1128/mSphere.00189-16.
- Wurster S, Bandi A, Beyda ND, et al. Drosophila melanogaster as a model to study virulence and azole treatment of the emerging pathogen Candida auris. J Antimicrob Chemother. 2019;74(7):1904–1910.
- Johnson CJ, Davis JM, Huttenlocher A, et al. Emerging fungal pathogen Candida auris evades neutrophil attack. mBio. 2018;9(4). DOI:10.1128/mBio.01403-18.
- Bruno M, Kersten S, Bain JM, et al. Transcriptional and functional insights into the host immune response against the emerging fungal pathogen Candida auris. Nat Microbiol. 2020;5(12):1516–1531.
- Potrykus J, Ballou ER, Childers DS, et al. Conflicting interests in the pathogen-host tug of war: fungal micronutrient scavenging versus mammalian nutritional immunity. PLoS Pathog. 2014;10(3):e1003910.
- Niemiec MJ, Grumaz C, Ermert D, et al. Dual transcriptome of the immediate neutrophil and Candida albicans interplay. BMC Genomics. 2017;18(1):696.
- Miramon P, Dunker C, Windecker H, et al. Cellular responses of Candida albicans to phagocytosis and the extracellular activities of neutrophils are critical to counteract carbohydrate starvation, oxidative and nitrosative stress. PLoS One. 2012;7(12):e52850.
- Miramon P, Kasper L, Hube B. Thriving within the host: candida spp. interactions with phagocytic cells. Med Microbiol Immunol. 2013;202(3):183–195.
- Duggan S, Essig F, Hünniger K, et al. Neutrophil activation by Candida glabrata but not Candida albicans promotes fungal uptake by monocytes. Cell Microbiol. 2015;17(9):1259–1276.
- Essig F, Hünniger K, Dietrich S, et al. Human neutrophils dump Candida glabrata after intracellular killing. Fungal Genet Biol. 2015;84:37–40.
- Campos-Garcia L, Jimenez-Valdes RJ, Hernandez-Bello R, et al. Candida albicans and non-albicans isolates from bloodstream have different capacities to induce neutrophil extracellular traps. J Fungi (Basel). 2019;5(2). DOI:10.3390/jof5020028.
- Willems HME, Stultz JS, Coltrane ME, et al. Disparate Candida albicans biofilm formation in clinical lipid emulsions due to capric acid-mediated inhibition. Antimicrob Agents Chemother. 2019;63(11). DOI:10.1128/AAC.01394-19.
- Glass KA, Longley SJ, Bliss JM, et al. Protection of Candida parapsilosis from neutrophil killing through internalization by human endothelial cells. Virulence. 2015;6(5):504–514.
- Cheng SC, Joosten LAB, Kullberg B-J, et al. Interplay between Candida albicans and the mammalian innate host defense. Infect Immun. 2012;80(4):1304–1313.
- Ramirez-Ortiz ZG, Means TK. The role of dendritic cells in the innate recognition of pathogenic fungi (A. fumigatus, C. neoformans and C. albicans). Virulence. 2012;3(7):635–646.
- Zipfel PF, Skerka C. Complement, Candida, and cytokines: the role of C5a in host response to fungi. Eur J Immunol. 2012;42(4):822–825.
- Rambach G, Speth C. Complement in Candida albicans infections. Front Biosci (Elite Ed). 2009;1:1–12.
- Cheng SC, Sprong T, Joosten LAB, et al. Complement plays a central role in Candida albicans-induced cytokine production by human PBMCs. Eur J Immunol. 2012;42(4):993–1004.
- Navarro-Arias MJ, Hernández-Chávez MJ, Garcia-Carnero LC, et al. Differential recognition of Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris by human innate immune cells. Infect Drug Resist. 2019;12:783–794.
- Kammer P, McNamara S, Wolf T, et al. Survival strategies of pathogenic Candida species in human blood show independent and specific adaptations. mBio. 2020;11(5). DOI:10.1128/mBio.02435-20.
- Hunniger K, Lehnert T, Bieber K, et al. A virtual infection model quantifies innate effector mechanisms and Candida albicans immune escape in human blood. PLoS Comput Biol. 2014;10(2):e1003479.
- Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5’-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198(2):179–182.
- Toth R, Alonso MF, Bain JM, et al. Different Candida parapsilosis clinical isolates and lipase deficient strain trigger an altered cellular immune response. Front Microbiol. 2015;6:1102.
- Pekmezovic M, Hovhannisyan H, Gresnigt MS, et al. Candida pathogens induce protective mitochondria-associated type I interferon signalling and a damage-driven response in vaginal epithelial cells. Nat Microbiol. 2021;6(5):643–657.
- Dunker C, Polke M, Schulze-Richter B, et al. Rapid proliferation due to better metabolic adaptation results in full virulence of a filament-deficient Candida albicans strain. Nat Commun. 2021;12(1):3899.
- Machata S, Sreekantapuram S, Hünniger K, et al. Significant differences in host-pathogen interactions between murine and human whole blood. Front Immunol. 2020;11:565869.
- Seelbinder B, et al. GEO2RNAseq: an easy-to-use R pipeline for complete pre-processing of RNA-seq data. biorxiv. 2019. https://doi.org/10.1101/771063.
- Edgar R, Domrachev M, Lash AE. Gene expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210.
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550.
- Howe KL, Achuthan P, Allen J, et al. Ensembl 2021. Nucleic Acids Res. 2021;49(D1):D884–D891.
- Team RC. R: a language and environment for statistical computing. Vienna Austria: R Foundation for Statistical Computing; 2021.
- Wickham H. ggplot2: elegant graphics for data analysis. second ed. New York: Springer Verlag; 2016.
- Casadevall A, Kontoyiannis DP, Robert V. On the emergence of Candida auris: climate change, azoles, swamps, and birds. mBio. 2019;10(4). DOI:10.1128/mBio.01397-19.
- Cuellar-Cruz M, López-Romero E, Ruiz-Baca E, et al. Differential response of Candida albicans and Candida glabrata to oxidative and nitrosative stresses. Curr Microbiol. 2014;69(5):733–739.
- Fradin C, De Groot P, MacCallum D, et al. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol. 2005;56(2):397–415.
- Naglik JR, Richardson JP, Moyes DL. Candida albicans pathogenicity and epithelial immunity. PLoS Pathog. 2014;10(8):e1004257.
- Kany S, Vollrath JT, Relja B. Cytokines in Inflammatory Disease. Int J Mol Sci. 2019;20(23):6008.
- He Y, Hara H, Nunez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci. 2016;41(12):1012–1021.
- Sokol CL, Luster AD. The chemokine system in innate immunity. Cold Spring Harb Perspect Biol. 2015;7(5):a016303.
- Lionakis MS, Fischer BG, Lim JK, et al. Chemokine receptor Ccr1 drives neutrophil-mediated kidney immunopathology and mortality in invasive candidiasis. PLoS Pathog. 2012;8(8):e1002865.
- Bai W, Wang Q, Deng Z, et al. TRAF1 suppresses antifungal immunity through CXCL1-mediated neutrophil recruitment during Candida albicans intradermal infection. Cell Commun Signal. 2020;18(1):30.
- Manicone AM, McGuire JK. Matrix metalloproteinases as modulators of inflammation. Semin Cell Dev Biol. 2008;19(1):34–41.
- Fingleton B. Matrix metalloproteinases as regulators of inflammatory processes. Biochim Biophys Acta Mol Cell Res. 2017;1864(11 Pt A):2036–2042.
- Hiyama A, Yokoyama K, Nukaga T, et al. Response to tumor necrosis factor-α mediated inflammation involving activation of prostaglandin E2 and Wnt signaling in nucleus pulposus cells. J Orthop Res. 2015;33(12):1756–1768.
- Ricciotti E, FitzGerald GA. Prostaglandins and inflammation. Arterioscler Thromb Vasc Biol. 2011;31(5):986–1000.
- Zittermann SI, Issekutz AC. Endothelial growth factors VEGF and bFGF differentially enhance monocyte and neutrophil recruitment to inflammation. J Leukoc Biol. 2006;80(2):247–257.
- Ma Y, Yang, X, Chatterjee, V, Meegan, JE, Beard, RS Jr, Yuan, SY, et al. Role of neutrophil extracellular traps and vesicles in regulating vascular endothelial permeability. Front Immunol. 2019 May 9;10:1037. doi:10.3389/fimmu.2019.01037.
- Sharma C, Kumar, N, Pandey, R, Meis, JF, Chowdhary, A, et al. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect. 2016 Jul 29;13:77–82. doi:10.1016/j.nmni.2016.07.003.
- Wirsching S, Michel, S, Köhler, G, Morschhäuser, J, et al. Activation of the multiple drug resistance gene MDR1 in fluconazole-resistant, clinical Candida albicans strains is caused by mutations in a trans-regulatory factor. J Bacteriol. 2000 Jan;182(2):400–404. doi:10.1128/JB.182.2.400-404.2000.
- Sun JN, Solis, NV, Phan, QT, Bajwa, JS, Kashleva, H, Thompson, A, Liu, Y, Dongari-Bagtzoglou, A, Edgerton, M, Filler, SG, et al. Host cell invasion and virulence mediated by Candida albicans Ssa1. PLoS Pathog. 2010 Nov 11;6(11):e1001181. doi:10.1371/journal.ppat.1001181.
- Altschul SF, Madden, TL, Schäffer, AA, Zhang, J, Zhang, Z, Miller, W, Lipman, DJ, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997 Sep 1;25(17):3389–3402. doi:10.1093/nar/25.17.3389.
- Dantas Ada S, Day A, Ikeh M, et al. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules. 2015;5(1):142–165.
- Cuellar-Cruz M, Briones-martin-del-campo M, Canas-Villamar I, et al. High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription Factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryot Cell. 2008;7(5):814–825.
- Gutierrez-Escobedo G, Hernández-Carreón O, Morales-Rojano B, et al. Candida glabrata peroxiredoxins, Tsa1 and Tsa2, and sulfiredoxin, Srx1, protect against oxidative damage and are necessary for virulence. Fungal Genet Biol. 2020;135:103287.
- Miramon P, Dunker, C, Kasper, L, Jacobsen, ID, Barz, D, Kurzai, O, Hube, B, et al. A family of glutathione peroxidases contributes to oxidative stress resistance in Candida albicans. Med Mycol. 2014 Apr;52(3):223–239. doi:10.1093/mmy/myt021.
- Shin DH, Jung S, Park SJ, et al. Characterization of thiol-specific antioxidant 1 (TSA1) of Candida albicans. Yeast. 2005;22(11):907–918.
- Wang Y, Cao YY, Jia XM, Cao YB, Gao PH, Fu XP, Ying K, Chen WS, Jiang YY Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic Biol Med. 2006;40(7):1201–1209. doi:10.1016/j.freeradbiomed.2005.11.019.
- Carreras MC, Pargament GA, Catz SD, et al. Kinetics of nitric oxide and hydrogen peroxide production and formation of peroxynitrite during the respiratory burst of human neutrophils. FEBS Lett. 1994;341(1):65–68.
- Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ . Invasive candidiasis. Nat Rev Dis Primers. 2018;4(1):18026. doi:10.1038/nrdp.2018.26.
- Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC . Hidden killers: human fungal infections. Sci Transl Med. 2012;4(165):165rv13. doi:10.1126/scitranslmed.3004404.
- Du H,Bing J, Hu T, Ennis CL, Nobile CJ, Huang G . Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020;16(10):e1008921 doi:10.1371/journal.ppat.1008921.
- Prestel C, Anderson E, Forsberg K, Lyman M, de Perio MA, Kuhar D, Edwards K, Rivera M, Shugart A, Walters M, Dotson NQ . Candida auris outbreak in a COVID-19 specialty care unit - Florida, July-August 2020. MMWR Morb Mortal Wkly Rep. 2021;70(2):56–57 doi:10.15585/mmwr.mm7002e3.
- Meis JF, Chowdhary A. Candida auris: a global fungal public health threat. Lancet Infect Dis. 2018;18(12):1298–1299.
- Ruiz-Gaitan A, Moret, AM, Tasias-Pitarch, M, Aleixandre-López, Al, Martínez-Morel, H, Calabuig, E, Salavert-Lletí, M, et al. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses. 2018;61(7):498–505. doi:10.1111/myc.12781.
- Ruiz-Gaitan A, Martínez, H, Moret, AM, Calabuig, E, Tasias, M, Alastruey-Izquierdo, A, Zaragoza, Ó, Mollar, J, et al. Detection and treatment of Candida auris in an outbreak situation: risk factors for developing colonization and candidemia by this new species in critically ill patients. Expert Rev Anti Infect Ther. 2019;17(4):295–305. doi:10.1080/14787210.2019.1592675.
- Desai JV, Lionakis MS. The role of neutrophils in host defense against invasive fungal infections. Curr Clin Microbiol Rep. 2018;5(3):181–189.
- Prausse MTE, Lehnert T, Timme S, et al. Predictive virtual infection modeling of fungal immune evasion in human whole blood. Front Immunol. 2018;9:560.
- Hunniger K, Bieber K, Martin R, et al. A second stimulus required for enhanced antifungal activity of human neutrophils in blood is provided by anaphylatoxin C5a. J Immunol. 2015;194(3):1199–1210.
- Fradin C, Kretschmar M, Nichterlein T, et al. Stage-specific gene expression of Candida albicans in human blood. Mol Microbiol. 2003;47(6):1523–1543.
- Grubb SE, Murdoch C, Sudbery PE, et al. Candida albicans-endothelial cell interactions: a key step in the pathogenesis of systemic Candidiasis. Infect Immun. 2008;76(10):4370–4377.
- Rubin-Bejerano I, Fraser I, Grisafi P, et al. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc Natl Acad Sci U S A. 2003;100(19):11007–11012.
- Garbe E, Vylkova S. Role of amino acid metabolism in the virulence of human pathogenic fungi. Curr Clin Micro Rep. 2019;6:108–119.
- Hornbach A, Heyken A, Schild L, et al. The Glycosylphosphatidylinositol-anchored protease SAP9 modulates the interaction of Candida albicans with human neutrophils. Infect Immun. 2009;77(12):5216–5224.
- Zawrotniak M, Bochenska O, Karkowska-Kuleta J, et al. aspartic proteases and major cell wall components in Candida albicans trigger the release of neutrophil extracellular traps. Front Cell Infect Microbiol. 2017;7:414.
- Rossato L, Colombo AL. Candida auris: What have we learned about its mechanisms of pathogenicity? Front Microbiol. 2018;9:3081.
- Cavalheiro M, Teixeira MC. Candida biofilms: threats, challenges, and promising strategies. Front Med (Lausanne). 2018;5:28.
- Biermann AR, Demers EG, Hogan DA. Mrr1 regulation of methylglyoxal catabolism and methylglyoxal-induced fluconazole resistance in Candida lusitaniae. Mol Microbiol. 2021;115(1):116–130.
- Demers EG, Stajich JE, Ashare A, et al. Balancing positive and negative selection: in vivo evolution of Candida lusitaniae MRR1. mBio. 2021;12(2). DOI:10.1128/mBio.03328-20.
- Alarco AM, Raymond M. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J Bacteriol. 1999;181(3):700–708.
- Sanguinetti M, Posteraro B, La Sorda M, et al. Role of AFR1, an ABC transporter-encoding gene, in the in vivo response to fluconazole and virulence of Cryptococcus neoformans. Infect Immun. 2006;74(2):1352–1359.
- Butler G, Rasmussen MD, Lin MF, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459(7247):657–662.
- Horton MV, Johnson CJ, Zarnowski R, et al. Candida auris cell wall mannosylation contributes to neutrophil evasion through pathways divergent from Candida albicans and Candida glabrata. mSphere. 2021;6(3):e0040621.
- Johnson CJ, Kernien JF, Hoyer AR, et al. Mechanisms involved in the triggering of neutrophil extracellular traps (NETs) by Candida glabrata during planktonic and biofilm growth. Sci Rep. 2017;7(1):13065.
- Linden JR, Maccani MA, Laforce-Nesbitt SS, et al. High efficiency opsonin-independent phagocytosis of Candida parapsilosis by human neutrophils. Med Mycol. 2010;48(2):355–364.
- Teixeira HD, Schumacher RI, Meneghini R. Lower intracellular hydrogen peroxide levels in cells overexpressing CuZn-superoxide dismutase. Proc Natl Acad Sci U S A. 1998;95(14):7872–7875.
- Cuéllar-Cruz M, Smietanka K, Minta Z, et al. Identification of Candida albicans heat shock proteins and Candida glabrata and Candida krusei enolases involved in the response to oxidative stress. Central European Journal of Biology. 2013;8(6):520–526.
- Gazendam RP, van Hamme JL, Tool ATJ, et al. Two independent killing mechanisms of Candida albicans by human neutrophils: evidence from innate immunity defects. Blood. 2014;124(4):590–597.
- Wellington M, Bliss JM, Haidaris CG. Enhanced phagocytosis of Candida species mediated by opsonization with a recombinant human antibody single-chain variable fragment. Infect Immun. 2003;71(12):7228–7231.
- Pereira HA, Hosking CS. The role of complement and antibody in opsonization and intracellular killing of Candida albicans. Clin Exp Immunol. 1984;57(2):307–314.
- Calvo B, Melo ASA, Perozo-Mena A, et al. First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J Infect. 2016;73(4):369–374.
- Chowdhary A, Sharma C, Duggal S, et al. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis. 2013;19(10):1670–1673.
- Hamal P, Kappe R, Rimek D. Rate of transmission and endogenous origin of Candida albicans and Candida glabrata on adult intensive care units studied by pulsed field gel electrophoresis. J Hosp Infect. 2001;49(1):37–42.
- Trofa D, Gacser A, Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev. 2008;21(4):606–625.
- Sanchez V, Vazquez JA, Barth-Jones D, et al. Nosocomial acquisition of Candida parapsilosis: an epidemiologic study. Am J Med. 1993;94(6):577–582.
- Pfaller MA. Nosocomial candidiasis: emerging species, reservoirs, and modes of transmission. Clin Infect Dis. 1996;22(2):S89–94.
- Sabino R, Sampaio P, Carneiro C, et al. Isolates from hospital environments are the most virulent of the Candida parapsilosis complex. BMC Microbiol. 2011;11(1):180.
- Piedrahita CT, Cadnum JL, Jencson AL, et al. Environmental surfaces in healthcare facilities are a potential source for transmission of Candida auris and other Candida species. Infect Control Hosp Epidemiol. 2017;38(9):1107–1109.
- Sabino R, Sampaio P, Rosado L, et al. Analysis of clinical and environmental Candida parapsilosis isolates by microsatellite genotyping–a tool for hospital infection surveillance. Clin Microbiol Infect. 2015;21(10):954 e1–8.
- Vallabhaneni S, Kallen A, Tsay S, et al. Investigation of the first seven reported cases of candida auris, a globally emerging invasive, multidrug-resistant fungus-United States, May 2013-August 2016. Am J Transplant. 2017;17(1):296–299.
- Ene IV, Cheng S-C, Netea MG, et al. Growth of Candida albicans cells on the physiologically relevant carbon source lactate affects their recognition and phagocytosis by immune cells. Infect Immun. 2013;81(1):238–248.
- Sherrington SL, Sorsby E, Mahtey N, et al. Adaptation of Candida albicans to environmental pH induces cell wall remodelling and enhances innate immune recognition. PLoS Pathog. 2017;13(5):e1006403.
- Odds FC, Van Nuffel L, Gow NAR. Survival in experimental Candida albicans infections depends on inoculum growth conditions as well as animal host. Microbiology (Reading). 2000;146(Pt 8):1881–1889.

