ABSTRACT
Mediator is a multisubunit complex conserved in eukaryotes that plays an essential coregulator role in RNA polymerase (Pol) II transcription. Despite intensive studies of the Mediator complex, the molecular mechanisms of its function in vivo remain to be fully defined. In this review, we will discuss the different aspects of Mediator function starting with its interactions with specific transcription factors, its recruitment to chromatin and how, as a coregulator, it contributes to the assembly of transcription machinery components within the preinitiation complex (PIC) in vivo and beyond the PIC formation.
Introduction
The discovery of Mediator originates from the observation of the activator interference phenomenon attributed to a competition between two activators for a common intermediate factor in yeast RNA polymerase II (Pol II) transcription in vitro.Citation1,2 These studies provided the first evidence for the existence of a Mediator of transcription regulation required for stimulation of transcription by activators. In parallel, genetic screens for suppressors of truncated C-terminal domain (CTD) of the largest Pol II subunit allowed to identify SRB (Suppressors of RNA polymerase B) genes. These genes corresponded to several Mediator subunits that interacted with Pol II CTD and were needed for activator-dependent transcription.Citation3-6 Multisubunit Mediator complex was then purified from yeast extracts. Mediator was shown to promote the assembly of general transcription factors (GTFs) TFIIA, B, D, E, F and H and Pol II into a preinitiation complex (PIC) on the promoter.Citation7,8 PIC formation represents a key intermediate in Pol II transcription and an important regulatory step. The Mediator complex was shown to enable activated transcription in vitro, but also to stimulate basal transcription in the absence of activator as well as Pol II CTD phosphorylation by Kin28/Cdk7 kinase of TFIIH.Citation9 Initial names for the yeast Mediator subunits derive from their identification in purified Mediator (Med) or genetic screens (Srb, Gal, Sin, Ssn, Nut, Rye). In human cells, complexes homologous to the yeast Mediator were purified via interactions with activation domains of different transcription factors, in particular nuclear receptors (NR).Citation10-17 Many names were initially used for human Mediator complexes depending on the purification procedures (TRAP, SMCC, CRSP, DRIP, ARC or PC2). Mediator has been then biochemically identified in many eukaryotes including Drosophila melanogaster and Caenorhabditis elegans, and more recently, in the plant Arabidopsis thaliana. A unified Mediator subunit nomenclature is now used in all eukaryotic organismsCitation18 (). Mediator is composed of 25 subunits in yeast and up to 30 subunits in human. Biochemical and structural studies revealed the modular organization of the complex. Four structural modules, head, middle, tail and Cdk8 kinase module, constitute the Mediator complex. The presence of a kinase module within the complex is transient. The spatial organization of Mediator modules that was recently redefined will be discussed below.
Table 1. Mediator subunits in yeast S. cerevisiae and human.Footnote*
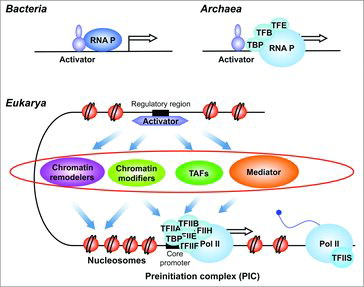
Multiple sequence alignments and analysis of secondary structure features led to the conclusion that Mediator was conserved in nearly all eukaryotes.Citation19 Metazoan Mediator can have up to five additional subunits to the 25 found in yeast. Together with other coregulator complexes, Mediator is characteristic of eukaryotic organisms and is not found in bacteria or Archaea. In eukaryotes, multisubunit coregulators constitute, thus, an additional regulatory layer between specific transcription factors and basal Pol II transcription machinery ().
Figure 1. Regulation of Pol II transcription initiation. A simplified view of eukaryotic Pol II transcription regulation at the step of preinitiation complex (PIC) formation is shown. Unlike bacteria which have direct links between activators and RNA polymerase or Archaea that have an RNA polymerase closely related to the eukaryotic enzyme and three general transcription factors, eukaryotes have multisubunit coregulators. These complexes constitute an additional regulatory layer (indicated by a red mark), that emerged in the eukaryotic kingdom, operating between activators and basal Pol II transcription machinery in chromatin context. These multisubunit complexes include chromatin remodelers and chromatin modifiers that act on the promoter chromatin structure or TAFs-containing TFIID and Mediator that stimulate PIC assembly. Some complexes could have both activities.
More than 20 years of intensive studies on the Mediator complex provided important information on its composition and function. General requirement of Mediator for Pol II recruitment and transcription has been demonstrated.Citation20-22 Mediator subunits were also implicated in human diseases including neurodevelopmental pathologies and different types of cancer.Citation23-26 In some cases, subunits of Mediator complex were proposed as potential therapeutic targets.Citation25,26 However, many open questions remain to be answered on the molecular mechanisms of Mediator function in vivo. In this review, we will discuss a current view of the molecular mechanisms in which Mediator is involved in vivo with a particular focus on its function in PIC assembly and raise some of the remaining open questions concerning its function in transcription regulation.
Mediator recruitment to chromatin and interactions with TFs
Mediator interactions with TFs
Mediator discovery has been conceptually associated with activated transcription in vitro. It is, thus, not surprising that several Mediator subunits interact with transcription activators. The Mediator tail module and, in particular, the Med2-Med3-Med15 (Gal11) submodule, was proposed to bind different activators since the deletion of some of Mediator subunits in yeast led to the loss of activator-dependent transcription response both in vitro and in vivo.Citation27-29 The fact that different Mediator subunits contact different TFs could, at least in part, reflect the differential involvement of Mediator subunits in regulation of a particular TF-dependent set of genes. The Med15 Mediator subunit in various species is involved in well-characterized interactions between Mediator and transcription activators. For example, NMR studies showed that a three-helix bundle “KIX” domain of the mammalian Med15 is engaged in Mediator interaction with SREBP-1a.Citation30 Med15 KIX domain is conserved in eukaryotes and, in fungi, is targeted by Oaf1 and Pdr1 activators.Citation31,32 Recently, the Med15 KIX domain — Pdr1 interaction was used to propose a target for antifungal strategies using molecules that disrupt the contact between the two proteins.Citation33 The KIX domain is not the only Med15 part that is implicated in contacts with activators. In yeast, Gcn4 binds Med15 through multiple low affinity interactions.Citation34,35 Structural analysis proposed a fuzzy protein interface between Gcn4 and Med15 in multiple conformations and orientations.Citation36,37 Med23, another tail module subunit specific to metazoan, has been initially identified in HeLa extracts as a target of adenovirus E1A protein.Citation10 This subunit was then shown to be required in mouse ES cells for proper PIC formation on gene promoters responsive to MAPK signaling TFs, E1A and Elk-1.Citation7,38 Interestingly, a recent work proposed a regulatory mechanism for Mediator-Elk-1 interaction through Med23 involving differential phosphorylation of Elk-1.Citation39
Mediator interactions with TFs are not restricted to the tail module. For example, the yeast Med17 Mediator head subunit interacts with Gal4 in vitroCitation40 and Drosophila kinase module subunits interact with TFs involved in Wnt signaling pathway.Citation41 In mammals, Med1 Mediator middle subunit is involved in a large number of interactions with NR.Citation10-17 Med1 binds an NR-activation domain through an LxxLL (Leucine XX Leucine Leucine) motif.Citation42 In addition, several studies have shown that Med14, linking tail, middle and head modules,Citation43-45 also interacts with different NRs.Citation46-48
Molecular mechanisms of transcription activation through Mediator-TF interactions remain to be understood and could potentially involve conformational changes in Mediator complex induced by TF binding.Citation49 It should be also noted that in many cases several TFs bind to the same gene promoter in vivo.Citation50 It would be important to know how these different regulatory signals are integrated by the Mediator complex and other coregulators recruited by TFs. One of the open questions is how the regulatory information is transmitted between different Mediator subunits within the complex.Citation51
Mediator chromatin binding
Direct binding of Mediator to DNA has not been documented. The Mediator complex is recruited to chromatin through its interactions with TFs, specific or general. Mediator enrichment can, thus, be expected both on regulatory regions, together with TFs, and on core promoter regions, where PIC components are located. Preferential association of yeast Mediator with upstream activating sequences of GAL genesCitation52 and Mediator recruitment to the promoter regions of constitutively expressed genesCitation53 were demonstrated by chromatin immunoprecipitation (ChIP). ChIP-chip genome-wide location analysis of different Mediator subunits in yeast revealed the presence of all four modules on the chromatin with a lower occupancy for the Cdk8 kinase module.Citation54 ChIP-seq approach allowed to gain in resolution for Mediator genome-wide distribution in yeastCitation55-58 and mammals.Citation59-61 Altogether, these studies demonstrated that Mediator associates essentially to the regulatory sequences with low signals on core promoters.Citation52-62 Mediator occupancy on transcribed regions was previously suggested, but several laboratories argued that Mediator ChIP signals on transcribed regions of highly expressed genes are non-specific.Citation55,56,63-66 Taking advantage of an independent technique, called chromatin endonuclease cleavage (ChEC)-seq, a recent study mapped the genome-wide yeast Mediator occupancy exclusively on the regulatory sequences.Citation67 Nevertheless, a transient Mediator association to core promoters was clearly evidenced by its stabilization within the PIC under conditions where Pol II CTD phosphorylation by TFIIH and therefore Pol II promoter escape were inhibited by depleting or inactivating Kin28 TFIIH kinase.Citation63,66 The model proposes that following Mediator recruitment to chromatin via Mediator-activator interactions on regulatory sequences, Mediator associates transiently with PIC components on core promoters to allow PIC formation. Mediator is then released upon CTD phosphorylation and Pol II promoter escape.
Important extension of these studies provided evidence and mechanistic clues to understand how Mediator composition evolves during transcription activation.Citation68,69 Mediator kinase module is known for its negative role in gene expression both in vitro and in vivo.Citation20,70-72 Its association with core Mediator is mutually exclusive with Pol II-Mediator interaction.Citation73-75 However, previous genome-wide location analysis of Mediator showed that the entire Mediator including kinase module is recruited to chromatin.Citation54 Based on the genome-wide occupancy of several Mediator subunits from all modules under Pol II CTD phosphorylation inhibition, recent studies proposed that the full Mediator complex is recruited to regulatory regions essentially through tail module interactions with TFs and then, kinase module is evicted to allow Mediator interactions with core promoter-bound PIC components. This mechanism involves changes in Mediator composition and emphasizes a regulatory function for the Mediator kinase module.Citation68,69
Contribution of structural studies to understanding Mediator mechanisms in PIC formation
Here, we will discuss how structural models of Mediator alone or in complex with Pol II contribute to our understanding of its role in PIC formation. Since the Cdk8 module represses PIC assembly, we will not discuss data that pertain to this module.
Mediator is large and flexible and not purified easily in a homogenous form. Hence, it is not readily amenable to the crystallographic investigations. The first studies resorted to electron microscopy (EM) to image negatively stained samples of Mediator devoid of the Cdk8 kinase module.Citation76-78 The general shape of free Mediator was apparent, being around 350 Å high and 200 Å wide and roughly triangular with a distinct dense domain at the base. Upon association with Pol II, Mediator assumed an elongated shape, embracing the enzyme. Tentative assignment of Mediator modules in the complex with Pol II was proposed. This interpretation formed the basis for nearly all the structural work that followed until it was revisited in depth 15 years later (see belowCitation79,80). Nevertheless, the early studies provided the important notion that Mediator could undergo large movements upon association with Pol II. They also established that the general structure of Mediator was similar in yeast, mouse and human. In addition, it was suggested that the interaction of Mediator with transcription activators could be the signal inducing the transition to an elongated shape allowing the interaction with Pol II.Citation81
Diffracting crystals could not be obtained for the entire Mediator complex. Nevertheless, the structures of several individual subunits expressed in vitro and then assembled in sub-complexes were resolved. Crystals of Saccharomyces cerevisiae and Schizosaccharomyces pombe head modules that diffracted at 4.3 Å and 3.4 Å, respectively, were obtained.Citation82,83 The general shape of the head module was compared with a crocodile head (with two jaws) and neck. Most of the carbon backbones of the individual subunits could be traced on the S. pombe head module. Mutations that affect the function of S. cerevisiae Mediator by impairing interactions with Pol II CTD,Citation6 its Rpb3 subunit,Citation21 or TFIIHCitation84 could be mapped on the head structure, suggesting the positions of interaction surfaces implicated in PIC formation.Citation83 These structures indicate that the head can adopt several alternative 3D organizations that may be implicated in the progressive assembly of the PIC. The atomic structure of complete Mediator middle module has not been determined. Nevertheless, the structures of two sub-complexes have been solved,Citation85,86 and completed by cross-linking data that were used to model the middle module.Citation87
Improvement of the biochemical purification of Mediator allowed for the analysis of its structure using cryo-EM.Citation79,80 Deletion mutants and tagging of Mediator subunits were used to investigate the position of the various subunits and modules. It was observed that Med14 subunit forms the backbone of Mediator, on which the head and middle modules associate. Med14 also connects to the tail. These findings were supported and expanded by an independent modeling of the yeast Mediator, based on integrative computational analysis, cross-linking data and available structural models.Citation44 These studies revisited the Mediator architecture and allowed for the reassignment of the modules both in isolated Mediator and in complex with Pol II. They also suggested intermediates in the formation of the Mediator-Pol II complex that may be relevant for PIC formation.
Recently, structures of Mediator alone or in complex with Pol II from S. pombe were further improved to 4.4 Å and 7.8 Å, respectively.Citation88 These structures included 16 out of the 21 subunits of the coregulator. This study further supports the notion that Med14, which extensively interacts with Med17, functions as a backbone on which the three modules are tethered. The formation of the Mediator-Pol II complex does not alter the conformation of Pol II contrary to that of Mediator in which the orientation of specific domains is altered due to Med14 change in conformation. These conformational changes allow the CTD to be engaged between the head and the middle modules and facilitate additional Mediator-Pol II contacts.Citation88 In yeast, the CTD consists of 25–26 heptad repeats. It could thus well interact when extended with several surfaces of Mediator and two other recent studies found that the CTD interacts with the head module.Citation45,89 All these works are in line with initial identification of Mediator by genetic studies indicating that its function is closely related to that of Pol II CTD.
Two recent studies have investigated the structure of the PIC and how Mediator participates in its organization.Citation45,90 A 15-subunit core Mediator composed of head and middle modules including Med14 was assembled on a core initially transcribing complex (cITC) of S. cerevisiae constituted of Pol II bound to an artificial transcription bubble together with TFIIB, TBP and TFIIF.Citation45 The structure of the Pol II-Mediator cITC has been solved at 9.7 ÅCitation45. Mediator-Pol II orientation proposed in S. cerevisaie cITC is similar to recently reported S. pombe Mediator-Pol II modelCitation88 leading to a consensus Mediator-Pol II conformation that is stable under cryo-EM conditions. However, other conformations of Mediator-Pol II that could be functional at different steps of PIC assembly may remain to be determined. In addition to gaining insights on the Mediator interaction with Pol II Rpb3–11 and Rpb4-Rpb7 subunits dimers, and Pol II dock, the study uncovered contacts with TFIIB. Mediator also participates in the stabilization of cITC. A lower resolution structure of a complete PIC assembled in vitro containing the 21 subunits Mediator, Pol II, TBP, TFIIA, TFIIB, TFIIE, TFIIF and TFIIH was also obtained from cryo-EM and cross-linking analysis.Citation90 It allowed to position all the components of the PIC and further supported the notion that the CTD-Mediator interface is the major contributor for the initial interaction of the coregulator and the enzyme.
Thus, structural information has proposed how Mediator functions to stimulate Pol II recruitment, to interact with CTD, to stabilize TFIIH and to interact with TFIIB. However, we are still far from an atomic understanding of the structure of Mediator or of the complete PIC. The preparations of the starting material (purified or recombinant) used in structural studies are different and it is likely that the assembly of the PIC entails several steps that might be captured by the different structures. One should also note that the models built from cryo-EM data are based on the selection of a limited number of images in the samples and represent only one possible conformation. In the future, we will need many more models representative of the alternative/successive steps leading to PIC assembly that will have to be supported by in vivo experiments. With this goal in mind, one should be able to relate the molecular function of Mediator in vitro with its role in PIC assembly on promoters in the chromatin context.
Mediator links with transcription machinery components in PIC assembly in vivo
Pioneering in vitro studies with purified PIC components led to the idea that the PIC is assembled starting with TFIID recruitment to promoter, followed by TFIIA and TFIIB incorporation, arrival of Pol II in association with TFIIF and final recruitment of TFIIH and TFIIE.Citation91,92 Mediator was absent from these initial experiments even though it is important for efficient PIC formation in vitro. Indeed, no stable PIC assembly on immobilized templates with nuclear extracts from yeast Mediator mutants is observed in its absence.Citation8 In a murine nuclear extract system, PIC assembly on promoter DNA in vitro was stimulated by activator–Mediator interactions.Citation7
To stabilize the PIC, Mediator cooperates with GTFs through physical protein–protein interactions or functional links. Specifically, cooperative action of Mediator and TFIID in PIC assembly was documented in vitro.Citation93-97 Human Mediator and TFIID are required for both basal and activated in vitro transcription in nuclear extracts.Citation93 In immobilized template assays reconstituted with purified proteins, human Mediator and TFIID assemble cooperatively on promoter DNA suggesting reciprocal and synergistic interactions between template DNA, activators, TFIID and Mediator required for activated transcription.Citation94 Using biochemical approaches, physical interactions between Mediator and TFIID were identified in yeastCitation95, Citation96 and human.Citation97 Based on in vitro experiments, a functional interplay between human Mediator and TFIIB was also suggestedCitation98 and interactions between yeast Mediator sub-complexes and TFIIB were shown.Citation99 Early Mediator studies demonstrated that in vitro Mediator stimulates the phosphorylation by TFIIH kinase of the Pol II CTD in yeast and mammalian cells,Citation9,100 emphasizing a functional link between Mediator and TFIIH complexes. As mentioned above, the discovery of Mediator complex is closely associated with its functional and direct interaction with Pol II.Citation4-6,9 These contacts were initially focused on the Pol II CTD and then extended to multiple subunits.Citation21,45,75,77,89,90,101-104
Many questions remain to be completely answered on the Mediator contribution to PIC assembly pathways in chromatin context and in gene-specific manner in vivo. Complementary to in vitro studies, Mediator function, in promoting PIC assembly in concert with the GTFs, was uncovered in vivo. In line with cooperative recruitment and action of Mediator and TFIID, TBP recruitment depends on Med17, the central head subunit. Indeed, med17–138 mutation led to a great decrease in TBP occupancy at several promoters.Citation105,106 Functional interactions between Drosophila Mediator and TFIID complexes were observed on metal-activated promoters.Citation107 More recently, an in vivo genome-wide study in Mediator Med17 mutants showed that Mediator selectively contributes to TBP recruitment or stabilization in the PIC.Citation56 Functional inteplay between Mediator and TFIIB was also proposed. Artificial tethering of TFIIB to the promoter led to Mediator recruitment and PIC assembly in vivo.Citation108 A recent genome-wide analysis of all PIC components occupancy showed that mutations in Med10 Mediator middle module that affect Mediator–TFIIB contact led to a pronounced decrease in TFIIB and had a global impact on Pol II recruitment and transcription.Citation57 This work demonstrated a role of Mediator in PIC assembly on a genomic scale at the step of TFIIB binding. Interestingly, the Mediator mutation effect on PIC assembly was related to the promoter architecture, suggesting a mechanism for functional interplay between Mediator and TFIIB in the context of promoter chromatin. A direct and functionally important link between Mediator and TFIIH core module was also identified.Citation56,84,109 An analysis of mutants in Med11 and Med17 Mediator head subunits showed that Mediator stabilizes TFIIK kinase and TFIIH core modules independently.Citation56,84 In line with close physical and functional associations between Mediator and Pol II, Mediator mutations or downregulation of Mediator subunits were shown to strongly compromise Pol II recruitment and transcription in yeast, mouse and human cells.Citation20,27,53,56,57,84,110-112 A direct Mediator-Pol II interaction was shown to be generally required for Pol II recruitment and transcription.Citation21 Using in vivo photo-cross-linking strategy, this work identified Rpb3 Pol II subunit that crosslinked to Med17 Mediator head module subunit among the 80 pairwise Pol II-Mediator contacts tested. The functional importance of this contact for the genome-wide recruitment of Pol II in PIC in vivo was demonstrated by complementary genetic and genomic approaches suggesting that most transcriptional regulatory information is transmitted through the Mediator-Pol II interface.
The PIC composition extends beyond the GTFs, Pol II and Mediator. For example, TFIIS, a thoroughly characterized transcription elongation factor, was identified as a PIC component by a proteomic analysis.Citation113 Mediator was found to act in conjunction with TFIIS in PIC assembly for optimal Pol II recruitment.Citation114 This conclusion is consistent with TFIIS contribution to in vitro PIC assembly independently from its role in stimulation of Pol II cleavage activity in transcription elongation.Citation113 Mediator cooperates also with other coactivators for optimal PIC assembly. Recent proteomic analysis identified Mediator and SAGA as major coactivator complexes within the PIC in HeLa and mouse ES cells.Citation115 Integration of the Mediator function with those of different chromatin regulators and other coactivator complexes would be important for our understanding of transcription regulation mechanisms.
The model for the PIC assembly in a linear sequence that does not include Mediator cannot explain in vivo observations suggesting an independent behavior for PIC components. Based on in vivo studies of Mediator mutants in yeast S. cerevisiae, a model for PIC assembly through multiple pathways was proposed.Citation84 Mediator was suggested to independently orchestrate multiple steps of PIC assembly in vivo.Citation56 Several studies support non-linear PIC assembly in vivo.Citation116 In addition, a partial PIC has been proposed as a regulated intermediate in response to heat shock in yeast.Citation117 Interestingly, a recent single-molecule analysis with human transcriptional platform reconstituted from purified TFIID, TFIIA and TFIIB revealed that promoter binding of TFIIB is highly transient and dynamic.Citation118 These works do not support the step-wise assembly model of GTFs in the PIC but rather suggest more complex and dynamic relationships between the PIC components.
Mediator function beyond the PIC assembly
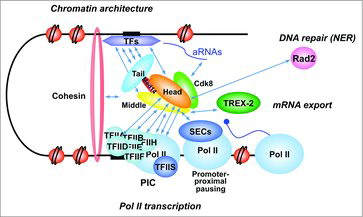
In vivo Mediator function in transcription regulation is not limited to promoting the efficient PIC assembly. The Mediator complex could also influence so-called post-recruitment steps of Pol II transcription and modulate the function and activity of the PIC after its formation. The best examples of such a role come from the Mediator studies in metazoan systems.Citation48,112,119,120 Mediator has been proposed to regulate promoter proximal pausing of Pol II, even though the exact mechanisms remain to be understood. For example, a model for Mediator function in Pol II transition to productive elongation has been proposed.Citation97 It has been suggested that metazoan-specific Med26 Mediator subunit can have a function helping to switch between transcription initiation to the elongation step by first contacting TFIID and then binding to the super-elongation complexes containing P-TEFb and other proteins.
A growing number of evidences suggest that in vivo Mediator function is closely related to chromatin architecture and that this complex can play an active role in 3D chromatin organization. In mouse cells, Mediator interacts with the cohesin complex in enhancer-core promoter loop formation that links very distant genome regions and activates transcription.Citation59 Mediator is highly enriched in groups of enhancers named super-enhancers important for regulation of lineage-specific genes.Citation60,61 In yeast, Mediator has been recently proposed as one of the complexes important for 3D genome organization.Citation121 A recent study suggests that Mediator can be implicated in gene positioning at the nuclear periphery through a direct contact with TREX-2 complex.Citation122 Other interesting works suggest that non-coding RNAs (aRNA and eRNA) could be involved in cooperative contacts with Mediator and other nuclear complexes in enhancer-core promoter looping that might represent an important mechanism for transcription regulation.Citation123,124
Concluding remarks and perspectives
Combined efforts taking advantage of complementarity between functional genome-wide, genetic, biochemical, structural and imaging approaches will continue to contribute to our understanding of complex mechanisms of Mediator function in transcription regulation. Integrative analyses and modeling will potentially constitute one of the challenges for the near future. It would be necessary to take into account multiple Mediator roles in vivo, from its contribution to the PIC assembly, to its function at post-recruitment steps of transcription, but also in chromatin organization and probably more generally in transcription-coupled events (). Specific Mediator subunits within the complex could be involved in different functions and a competition between different proteins to the same interaction interface on the Mediator complex could not be excluded. Future studies are needed to precisely define interaction interfaces involved. In vitro studiesCitation125 and a recent work based on single-cell imagingCitation126 suggested also an active role for Mediator in re-initiation of transcription, but this phenomenon has not been observed in vivo. Mediator functions beyond transcription are emerging suggesting that Mediator might act as an assembly platform or a regulatory element in chromatin-related processes including DNA repairCitation55 or telomere maintenance.Citation127,128 The modular Mediator organization is central for its functions and the precise mechanisms of Cdk8 kinase module remain to be fully understood. It would be particularly interesting to precisely define how individual Mediator subunits contribute to the overall function of the complex. For the moment, in vivo Mediator complex assembly and its regulation remain completely unknown. The yeast model will continue to offer a lot of possibilities to study the molecular mechanisms of Mediator complex that are likely conserved in other eukaryotes including human. Studies in mammalian systems are essential to understand Mediator implications in differentiation, cell type-specific transcription programs and human diseases. In this regard, the focus on metazoan-specific Mediator subunits would be of particular interest.
Figure 2. Mediator interactions within the nucleus. Mediator functions are closely related to its physical and functional interactions with nuclear proteins. Some of these contacts discussed in the review are summarized on the figure focusing on Pol II transcription (TF interactions, PIC assembly, promoter proximal pausing), but also extending to chromatin architecture, mRNA export or DNA repair. The cartoon represents a combined view from different studies in yeasts and metazoans. Identified interactions between Mediator and nuclear proteins and ncRNAs are shown. Mediator contacts with SECs, cohesin and ncRNAs were reported in metazoans. Mediator modules are shown in different colors with Med14 linking the three main modules (head, middle and tail).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Agence Nationale de la Recherche under Grant n° ANR-14-CE10–0012–01; and the Fondation ARC under Grant n SL220130607079. T.E was supported by a grant from the Fondation pour la Recherche Médicale (FDT20150531985).
References
- Flanagan PM, Kelleher RJ, Sayre MH, Tschochner H, Kornberg RD. A mediator required for activation of RNA polymerase II transcription in vitro. Nature 1991; 350(6317):436-438; PMID:2011193; https://doi.org/10.1038/350436a0
- Kelleher RJ, 3rd, Flanagan PM, Kornberg RD. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 1990; 61(7):1209-1215; PMID:2163759; https://doi.org/10.1016/0092-8674(90)90685-8
- Hengartner CJ, Thompson CM, Zhang J, Chao DM, Liao SM, Koleske AJ, Okamura S, Young RA. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev 1995; 9(8):897-910; PMID:7774808; https://doi.org/10.1101/gad.9.8.897
- Koleske AJ, Young RA. An RNA polymerase II holoenzyme responsive to activators. Nature 1994; 368(6470):466-469; PMID:8133894; https://doi.org/10.1038/368466a0
- Nonet ML, Young RA. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics 1989; 123(4):715-724; PMID:2693207
- Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 1993; 73(7):1361-1375; PMID:8324825; https://doi.org/10.1016/0092-8674(93)90362-T
- Cantin GT, Stevens JL, Berk AJ. Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc Natl Acad Sci USA 2003; 100(21):12003-12008; PMID:14506297; https://doi.org/10.1073/pnas.2035253100
- Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev 1999; 13(1):49-63; PMID:9887099; https://doi.org/10.1101/gad.13.1.49
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 1994; 77(4):599-608; PMID:8187178; https://doi.org/10.1016/0092-8674(94)90221-6
- Boyer TG, Martin ME, Lees E, Ricciardi RP, Berk AJ. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 1999; 399(6733):276-279; PMID:10353252; https://doi.org/10.1038/20466
- Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA 1996; 93(16):8329-8333; PMID:8710870; https://doi.org/10.1073/pnas.93.16.8329
- Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell 1999; 3(1):97-108; PMID:10024883; https://doi.org/10.1016/S1097-2765(00)80178-1
- Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell 1999; 3(3):361-370; PMID:10198638; https://doi.org/10.1016/S1097-2765(00)80463-3
- Naar AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 1999; 398(6730):828-832; PMID:10235267; https://doi.org/10.1038/19789
- Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Näär AM, Erdjument-Bromage H, Tempst P, Freedman LP. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 1999; 398(6730):824-828; PMID:10235266; https://doi.org/10.1038/19783
- Ryu S, Zhou S, Ladurner AG, Tjian R. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 1999; 397(6718):446-450; PMID:9989412; https://doi.org/10.1038/17141
- Malik S, Gu W, Wu W, Qin J, Roeder RG. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol Cell 2000; 5(4):753-760; PMID:10882111; https://doi.org/10.1016/S1097-2765(00)80254-3
- Bourbon HM, Aguilera A, Ansari AZ, Asturias FJ, Berk AJ, Bjorklund S, Blackwell TK, Borggrefe T, Carey M, Carlson M et al. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell 2004; 14(5):553-557; PMID:15175151; https://doi.org/10.1016/j.molcel.2004.05.011
- Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res 2008; 36(12):3993-4008; PMID:18515835; https://doi.org/10.1093/nar/gkn349
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 1998; 95(5):717-728; PMID:9845373; https://doi.org/10.1016/S0092-8674(00)81641-4
- Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science 2011; 331(6023):1451-1454; PMID:21415355; https://doi.org/10.1126/science.1200188
- Thompson CM, Young RA. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA 1995; 92(10):4587-4590; PMID:7753848; https://doi.org/10.1073/pnas.92.10.4587
- Hashimoto S, Boissel S, Zarhrate M, Rio M, Munnich A, Egly JM, Colleaux L. MED23 mutation links intellectual disability to dysregulation of immediate early gene expression. Science 2011; 333(6046):1161-1163; PMID:21868677; https://doi.org/10.1126/science.1206638
- Kaufmann R, Straussberg R, Mandel H, Fattal-Valevski A, Ben-Zeev B, Naamati A, Shaag A, Zenvirt S, Konen O, Mimouni-Bloch A et al. Infantile cerebral and cerebellar atrophy is associated with a mutation in the MED17 subunit of the transcription preinitiation mediator complex. Am J Hum Genet 2010; 87(5):667-670; PMID:20950787; https://doi.org/10.1016/j.ajhg.2010.09.016
- Schiano C, Casamassimi A, Rienzo M, de Nigris F, Sommese L, Napoli C. Involvement of Mediator complex in malignancy. Biochim Biophys Acta 2014; 1845(1):66-83; PMID:24342527; https://doi.org/10.1016/j.bbcan.2013.12.001
- Spaeth JM, Kim NH, Boyer TG. Mediator and human disease. Semin Cell Dev Biol 2011; 22(7):776-787; PMID:21840410; https://doi.org/10.1016/j.semcdb.2011.07.024
- Han SJ, Lee YC, Gim BS, Ryu GH, Park SJ, Lane WS, Kim YJ. Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation. Mol Cell Biol 1999; 19(2):979-988; PMID:9891034; https://doi.org/10.1128/MCB.19.2.979
- Lee YC, Park JM, Min S, Han SJ, Kim YJ. An activator binding module of yeast RNA polymerase II holoenzyme. Mol Cell Biol 1999; 19(4):2967-2976; PMID:10082564; https://doi.org/10.1128/MCB.19.4.2967
- Myers LC, Gustafsson CM, Hayashibara KC, Brown PO, Kornberg RD. Mediator protein mutations that selectively abolish activated transcription. Proc Natl Acad Sci USA 1999; 96(1):67-72; PMID:9874773; https://doi.org/10.1073/pnas.96.1.67
- Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 2006; 442(7103):700-704; PMID:16799563; https://doi.org/10.1038/nature04942
- Thakur JK, Arthanari H, Yang F, Chau KH, Wagner G, Näär AM. Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J Biol Chem 2009; 284(7):4422-4428; PMID:19056732; https://doi.org/10.1074/jbc.M808263200
- Thakur JK, Arthanari H, Yang F, Pan SJ, Fan X, Breger J, Frueh DP, Gulshan K, Li DK, Mylonakis E et al. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 2008; 452(7187):604-609; PMID:18385733; https://doi.org/10.1038/nature06836
- Nishikawa JL, Boeszoermenyi A, Vale-Silva LA, Torelli R, Posteraro B, Sohn YJ, Ji F, Gelev V, Sanglard D, Sanguinetti M et al. Inhibiting fungal multidrug resistance by disrupting an activator-Mediator interaction. Nature 2016; 530(7591):485-489; PMID:26886795; https://doi.org/10.1038/nature16963
- Herbig E, Warfield L, Fish L, Fishburn J, Knutson BA, Moorefield B, Pacheco D, Hahn S. Mechanism of Mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains. Mol Cell Biol 2010; 30(10):2376-2390; PMID:20308326; https://doi.org/10.1128/MCB.01046-09
- Jedidi I, Zhang F, Qiu H, Stahl SJ, Palmer I, Kaufman JD, Nadaud PS, Mukherjee S, Wingfield PT, Jaroniec CP et al. Activator Gcn4 employs multiple segments of Med15/Gal11, including the KIX domain, to recruit mediator to target genes in vivo. J Biol Chem 2010; 285(4):2438-2455; PMID:19940160; https://doi.org/10.1074/jbc.M109.071589
- Brzovic PS, Heikaus CC, Kisselev L, Vernon R, Herbig E, Pacheco D, Warfield L, Littlefield P, Baker D, Klevit RE et al. The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol Cell 2011; 44(6):942-953; PMID:22195967; https://doi.org/10.1016/j.molcel.2011.11.008
- Warfield L, Tuttle LM, Pacheco D, Klevit RE, Hahn S. A sequence-specific transcription activator motif and powerful synthetic variants that bind Mediator using a fuzzy protein interface. Proc Natl Acad Sci USA 2014; 111(34):E3506-3513; PMID:25122681; https://doi.org/10.1073/pnas.1412088111
- Stevens JL, Cantin GT, Wang G, Shevchenko A, Shevchenko A, Berk AJ. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 2002; 296(5568):755-758; PMID:11934987; https://doi.org/10.1126/science.1068943
- Mylona A, Theillet FX, Foster C, Cheng TM, Miralles F, Bates PA, Selenko P, Treisman R. Opposing effects of Elk-1 multisite phosphorylation shape its response to ERK activation. Science 2016; 354(6309):233-237; PMID:27738173; https://doi.org/10.1126/science.aad1872
- Koh SS, Ansari AZ, Ptashne M, Young RA. An activator target in the RNA polymerase II holoenzyme. Mol Cell 1998; 1(6):895-904; PMID:9660972; https://doi.org/10.1016/S1097-2765(00)80088-X
- Carrera I, Janody F, Leeds N, Duveau F, Treisman JE. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc Natl Acad Sci USA 2008; 105(18):6644-6649; PMID:18451032; https://doi.org/10.1073/pnas.0709749105
- Malik S, Guermah M, Yuan CX, Wu W, Yamamura S, Roeder RG. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol Cell Biol 2004; 24(18):8244-8254; PMID:15340084; https://doi.org/10.1128/MCB.24.18.8244-8254.2004
- Cevher MA, Shi Y, Li D, Chait BT, Malik S, Roeder RG. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat Struct Mol Biol 2014; 21(12):1028-1034; PMID:25383669; https://doi.org/10.1038/nsmb.2914
- Robinson PJ, Trnka MJ, Pellarin R, Greenberg CH, Bushnell DA, Davis R, Burlingame AL, Sali A, Kornberg RD. Molecular architecture of the yeast Mediator complex. Elife 2015; 4; https://doi.org/10.7554/eLife.08719
- Plaschka C, Larivière L, Wenzeck L, Seizl M, Hemann M, Tegunov D, Petrotchenko EV, Borchers CH, Baumeister W, Herzog F et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 2015; 518(7539):376-380; PMID:25652824; https://doi.org/10.1038/nature14229
- Grontved L, Madsen MS, Boergesen M, Roeder RG, Mandrup S. MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor gamma and is required for full transcriptional activity and adipogenesis. Mol Cell Biol 2010; 30(9):2155-2169; PMID:20194623; https://doi.org/10.1128/MCB.01238-09
- Hittelman AB, Burakov D, Iniguez-Lluhi JA, Freedman LP, Garabedian MJ. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J 1999; 18(19):5380-5388; PMID:10508170; https://doi.org/10.1093/emboj/18.19.5380
- Malik S, Wallberg AE, Kang YK, Roeder RG. TRAP/SMCC/mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol Cell Biol 2002; 22(15):5626-5637; PMID:12101254; https://doi.org/10.1128/MCB.22.15.5626-5637.2002
- Taatjes DJ, Naar AM, Andel F, 3rd, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 2002; 295(5557):1058-1062; PMID:11834832; https://doi.org/10.1126/science.1065249
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J et al. Transcriptional regulatory code of a eukaryotic genome. Nature 2004; 431(7004):99-104; PMID:15343339; https://doi.org/10.1038/nature02800
- Niederberger T, Etzold S, Lidschreiber M, Maier KC, Martin DE, Fröhlich H, Cramer P, Tresch A. MC EMiNEM maps the interaction landscape of the Mediator. PLoS Comput Biol 2012; 8(6):e1002568; PMID:22737066; https://doi.org/10.1371/journal.pcbi.1002568
- Kuras L, Borggrefe T, Kornberg RD. Association of the Mediator complex with enhancers of active genes. Proc Natl Acad Sci USA 2003; 100(24):13887-13891; PMID:14623974; https://doi.org/10.1073/pnas.2036346100
- Ansari SA, He Q, Morse RH. Mediator complex association with constitutively transcribed genes in yeast. Proc Natl Acad Sci USA 2009; 106(39):16734-16739; PMID:19805365; https://doi.org/10.1073/pnas.0905103106
- Andrau JC, van de Pasch L, Lijnzaad P, Bijma T, Koerkamp MG, van de Peppel J, Werner M, Holstege FC. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol Cell 2006; 22(2):179-192; PMID:16630888; https://doi.org/10.1016/j.molcel.2006.03.023
- Eyboulet F, Cibot C, Eychenne T, Neil H, Alibert O, Werner M, Soutourina J. Mediator links transcription and DNA repair by facilitating Rad2/XPG recruitment. Genes Dev 2013; 27(23):2549-2562; PMID:24298055; https://doi.org/10.1101/gad.225813.113
- Eyboulet F, Wydau-Dematteis S, Eychenne T, Alibert O, Neil H, Boschiero C, Nevers MC, Volland H, Cornu D, Redeker V. Mediator independently orchestrates multiple steps of preinitiation complex assembly in vivo. Nucleic Acids Res 2015; 43(19):9214-9231; PMID:26240385; https://doi.org/10.1093/nar/gkv782
- Eychenne T, Novikova E, Barrault MB, Alibert O, Boschiero C, Peixeiro N, Cornu D, Redeker V, Kuras L, Nicolas P et al. Functional interplay between Mediator and TFIIB in preinitiation complex assembly in relation to promoter architecture. Genes Dev 2016; 30(18):2119-2132; PMID:27688401; https://doi.org/10.1101/gad.285775.116
- Wong KH, Struhl K. The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev 2011; 25(23):2525-2539; PMID:22156212; https://doi.org/10.1101/gad.179275.111
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010; 467(7314):430-435; PMID:20720539; https://doi.org/10.1038/nature09380
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153(2):320-334; PMID:23582323; https://doi.org/10.1016/j.cell.2013.03.036
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153(2):307-319; PMID:23582322; https://doi.org/10.1016/j.cell.2013.03.035
- Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell 2011; 41(4):480-492; PMID:21329885; https://doi.org/10.1016/j.molcel.2011.01.015
- Jeronimo C, Robert F. Kin28 regulates the transient association of Mediator with core promoters. Nat Struct Mol Biol 2014; 21(5):449-455; PMID:24704787; https://doi.org/10.1038/nsmb.2810
- Park D, Lee Y, Bhupindersingh G, Iyer VR. Widespread misinterpretable ChIP-seq bias in yeast. PLoS One 2013; 8(12):e83506; PMID:24349523; https://doi.org/10.1371/journal.pone.0083506
- Teytelman L, Thurtle DM, Rine J, van Oudenaarden A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci USA 2013; 110(46):8602-18607; PMID:24173036; https://doi.org/10.1073/pnas.1316064110
- Wong KH, Jin Y, Struhl K. TFIIH Phosphorylation of the Pol II CTD stimulates mediator dissociation from the Preinitiation complex and promoter escape. Mol Cell 2014; 54(4):601-612; PMID:24746699; https://doi.org/10.1016/j.molcel.2014.03.024
- Grunberg S, Henikoff S, Hahn S, Zentner GE. Mediator binding to UASs is broadly uncoupled from transcription and cooperative with TFIID recruitment to promoters. EMBO J 2016; 35(22):2435-2446; PMID:27797823; https://doi.org/10.15252/embj.201695020
- Jeronimo C, Langelier MF, Bataille AR, Pascal JM, Pugh BF, Robert F. Tail and Kinase modules differently regulate core mediator recruitment and function in vivo. Mol Cell 2016; 64(3):455-466; PMID:27773677; https://doi.org/10.1016/j.molcel.2016.09.002
- Petrenko N, Jin Y, Wong KH, Struhl K. Mediator undergoes a compositional change during transcriptional activation. Mol Cell 2016; 64(3):443-454; PMID:27773675; https://doi.org/10.1016/j.molcel.2016.09.015
- Kemmeren P, Sameith K, van de Pasch LA, Benschop JJ, Lenstra TL, Margaritis T, O'Duibhir E, Apweiler E, van Wageningen S, Ko CW et al. Large-scale genetic perturbations reveal regulatory networks and an abundance of gene-specific repressors. Cell 2014; 157(3):740-752; PMID:24766815; https://doi.org/10.1016/j.cell.2014.02.054
- Myers LC, Gustafsson CM, Bushnell DA, Lui M, Erdjument-Bromage H, Tempst P, Kornberg RD. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev 1998; 12(1):45-54; PMID:9420330; https://doi.org/10.1101/gad.12.1.45
- van de Peppel J, Kettelarij N, van Bakel H, Kockelkorn TT, van Leenen D, Holstege FC. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol Cell 2005; 19(4):511-522; PMID:16109375; https://doi.org/10.1016/j.molcel.2005.06.033
- Naar AM, Taatjes DJ, Zhai W, Nogales E, Tjian R. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev 2002; 16(11):1339-1344; PMID:12050112; https://doi.org/10.1101/gad.987602
- Spahr H, Khorosjutina O, Baraznenok V, Linder T, Samuelsen CO, Hermand D, Mäkela TP, Holmberg S, Gustafsson CM. Mediator influences Schizosaccharomyces pombe RNA polymerase II-dependent transcription in vitro. J Biol Chem 2003; 278(51):51301-51306; PMID:14534314; https://doi.org/10.1074/jbc.M306750200
- Tsai KL, Sato S, Tomomori-Sato C, Conaway RC, Conaway JW, Asturias FJ. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat Struct Mol Biol 2013; 20(5):611-619; PMID:23563140; https://doi.org/10.1038/nsmb.2549
- Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. Conserved structures of mediator and RNA polymerase II holoenzyme. Science 1999; 283(5404):985-987; PMID:9974391; https://doi.org/10.1126/science.283.5404.985
- Davis JA, Takagi Y, Kornberg RD, Asturias FA. Structure of the yeast RNA polymerase II holoenzyme: mediator conformation and polymerase interaction. Mol Cell 2002; 10(2):409-415; PMID:12191485; https://doi.org/10.1016/S1097-2765(02)00598-1
- Dotson MR, Yuan CX, Roeder RG, Myers LC, Gustafsson CM, Jiang YW, Li Y, Kornberg RD, Asturias FJ. Structural organization of yeast and mammalian mediator complexes. Proc Natl Acad Sci USA 2000; 97(26):14307-14310; PMID:11114191; https://doi.org/10.1073/pnas.260489497
- Tsai KL, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 2014; 157(6):1430-1444; PMID:24882805; https://doi.org/10.1016/j.cell.2014.05.015
- Wang X, Sun Q, Ding Z, Ji J, Wang J, Kong X, Yang J, Cai G. Redefining the modular organization of the core Mediator complex. Cell Res 2014; 24(7):796-808; PMID:24810298; https://doi.org/10.1038/cr.2014.64
- Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol 2013; 48(6):575-608; PMID:24088064; https://doi.org/10.3109/10409238.2013.840259
- Imasaki T, Calero G, Cai G, Tsai KL, Yamada K, Cardelli F, Erdjument-Bromage H, Tempst P, Berger I, Kornberg GL. Architecture of the mediator head module. Nature 2011; 475(7355):240-243; PMID:21725323; https://doi.org/10.1038/nature10162
- Lariviere L, Plaschka C, Seizl M, Wenzeck L, Kurth F, Cramer P. Structure of the Mediator head module. Nature 2012; 492(7429):448-451; PMID:23123849; https://doi.org/10.1038/nature11670
- Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell 2008; 31(3):337-346; PMID:18691966; https://doi.org/10.1016/j.molcel.2008.06.021
- Baumli S, Hoeppner S, Cramer P. A conserved mediator hinge revealed in the structure of the MED7.MED21 (Med7.Srb7) heterodimer. J Biol Chem 2005; 280(18):18171-18178; PMID:15710619; https://doi.org/10.1074/jbc.M413466200
- Koschubs T, Seizl M, Larivière L, Kurth F, Baumli S, Martin DE, Cramer P. Identification, structure, and functional requirement of the Mediator submodule Med7N/31. EMBO J 2009; 28(1):69-80; PMID:19057509; https://doi.org/10.1038/emboj.2008.254
- Lariviere L, Yu X, Gopalan S, Chao TC, Zhang Y, Florens L, Washburn MP, Murakami K, Conaway RC, Conaway JW et al. Model of the Mediator middle module based on protein cross-linking. Nucleic Acids Res 2013; 41(20):9266-9273; PMID:23939621; https://doi.org/10.1093/nar/gkt704
- Tsai KL, Yu X, Gopalan S, Chao TC, Zhang Y, Florens L, Washburn MP, Murakami K, Conaway RC, Conaway JW et al. Mediator structure and rearrangements required for holoenzyme formation. Nature 2017; 544(7649):196-201; PMID:28241144; https://doi.org/10.1038/nature21393
- Robinson PJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD. Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proc Natl Acad Sci USA 2012; 109(44):17931-17935; PMID:23071300; https://doi.org/10.1073/pnas.1215241109
- Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei PJ, Burlingame AL, Kornberg RD. Structure of a complete Mediator-RNA Polymerase II Pre-initiation complex. Cell 2016; 166(6):1411-1422 e1416; PMID:27610567; https://doi.org/10.1016/j.cell.2016.08.050
- Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 1989; 56(4):549-561; PMID:2917366; https://doi.org/10.1016/0092-8674(89)90578-3
- Ranish JA, Hahn S. Transcription: basal factors and activation. Curr Opin Genet Dev 1996; 6(2):151-158; PMID:8722170; https://doi.org/10.1016/S0959-437X(96)80044-X
- Baek HJ, Malik S, Qin J, Roeder RG. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol Cell Biol 2002; 22(8):2842-2852; PMID:11909976; https://doi.org/10.1128/MCB.22.8.2842-2852.2002
- Johnson KM, Wang J, Smallwood A, Arayata C, Carey M. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev 2002; 16(14):1852-1863; PMID:12130544; https://doi.org/10.1101/gad.995702
- Koleske AJ, Buratowski S, Nonet M, Young RA. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell 1992; 69(5):883-894; PMID:1591782; https://doi.org/10.1016/0092-8674(92)90298-Q
- Lariviere L, Geiger S, Hoeppner S, Röther S, Strässer K, Cramer P. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat Struct Mol Biol 2006; 13(10):895-901; PMID:16964259; https://doi.org/10.1038/nsmb1143
- Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 2011; 146(1):92-104; PMID:21729782; https://doi.org/10.1016/j.cell.2011.06.005
- Baek HJ, Kang YK, Roeder RG. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol Chem 2006; 281(22):15172-15181; PMID:16595664; https://doi.org/10.1074/jbc.M601983200
- Kang JS, Kim SH, Hwang MS, Han SJ, Lee YC, Kim YJ. The structural and functional organization of the yeast mediator complex. J Biol Chem 2001; 276(45):42003-42010; PMID:11555651; https://doi.org/10.1074/jbc.M105961200
- Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci USA 1998; 95(15):8538-8543; PMID:9671713; https://doi.org/10.1073/pnas.95.15.8538
- Cai G, Imasaki T, Takagi Y, Asturias FJ. Mediator structural conservation and implications for the regulation mechanism. Structure 2009; 17(4):559-567; PMID:19368889; https://doi.org/10.1016/j.str.2009.01.016
- Cai G, Imasaki T, Yamada K, Cardelli F, Takagi Y, Asturias FJ. Mediator head module structure and functional interactions. Nat Struct Mol Biol 2010; 17(3):273-279; PMID:20154708; https://doi.org/10.1038/nsmb.1757
- Reeves WM, Hahn S. Activator-independent functions of the yeast mediator sin4 complex in preinitiation complex formation and transcription reinitiation. Mol Cell Biol 2003; 23(1):349-358; PMID:12482986; https://doi.org/10.1128/MCB.23.1.349-358.2003
- Tardiff DF, Abruzzi KC, Rosbash M. Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proc Natl Acad Sci USA 2007; 104(50):19948-19953; PMID:18077427; https://doi.org/10.1073/pnas.0710179104
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 1999; 399(6736):609-613; PMID:10376605; https://doi.org/10.1038/21239
- Li XY, Virbasius A, Zhu X, Green MR. Enhancement of TBP binding by activators and general transcription factors. Nature 1999; 399(6736):605-609; PMID:10376604; https://doi.org/10.1038/21232
- Marr MT, 2nd, Isogai Y, Wright KJ, Tjian R. Coactivator cross-talk specifies transcriptional output. Genes Dev 2006; 20(11):1458-1469; PMID:16751183; https://doi.org/10.1101/gad.1418806
- Lacombe T, Poh SL, Barbey R, Kuras L. Mediator is an intrinsic component of the basal RNA polymerase II machinery in vivo. Nucleic Acids Res 2013; 41(21):9651-9662; PMID:23963697; https://doi.org/10.1093/nar/gkt701
- Seizl M, Lariviere L, Pfaffeneder T, Wenzeck L, Cramer P. Mediator head subcomplex Med11/22 contains a common helix bundle building block with a specific function in transcription initiation complex stabilization. Nucleic Acids Res 2011; 39(14):6291-6304; PMID:21498544; https://doi.org/10.1093/nar/gkr229
- Qiu H, Hu C, Yoon S, Natarajan K, Swanson MJ, Hinnebusch AG. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol Cell Biol 2004; 24(10):4104-4117; PMID:15121833; https://doi.org/10.1128/MCB.24.10.4104-4117.2004
- Rana R, Surapureddi S, Kam W, Ferguson S, Goldstein JA. Med25 is required for RNA polymerase II recruitment to specific promoters, thus regulating xenobiotic and lipid metabolism in human liver. Mol Cell Biol 2011; 31(3):466-481; PMID:21135126; https://doi.org/10.1128/MCB.00847-10
- Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell 2005; 17(5):683-694; PMID:15749018; https://doi.org/10.1016/j.molcel.2005.02.010
- Kim B, Nesvizhskii AI, Rani PG, Hahn S, Aebersold R, Ranish JA. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc Natl Acad Sci USA 2007; 104(41):16068-16073; PMID:17913884; https://doi.org/10.1073/pnas.0704573104
- Guglielmi B, Soutourina J, Esnault C, Werner M. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc Natl Acad Sci USA 2007; 104(41):16062-16067; PMID:17901206; https://doi.org/10.1073/pnas.0704534104
- Chen XF, Lehmann L, Lin JJ, Vashisht A, Schmidt R, Ferrari R, Huang C, McKee R, Mosley A, Plath K et al. Mediator and SAGA have distinct roles in Pol II preinitiation complex assembly and function. Cell Rep 2012; 2(5):1061-1067; PMID:23177621; https://doi.org/10.1016/j.celrep.2012.10.019
- Sikorski TW, Buratowski S. The basal initiation machinery: beyond the general transcription factors. Curr Opin Cell Biol 2009; 21(3):344-351; PMID:19411170; https://doi.org/10.1016/j.ceb.2009.03.006
- Zanton SJ, Pugh BF. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev 2006; 20(16):2250-2265; PMID:16912275; https://doi.org/10.1101/gad.1437506
- Zhang Z, English BP, Grimm JB, Kazane SA, Hu W, Tsai A, Inouye C, You C, Piehler J, Schultz PG et al. Rapid dynamics of general transcription factor TFIIB binding during preinitiation complex assembly revealed by single-molecule analysis. Genes Dev 2016; 30(18):2106-2118; PMID:27798851; https://doi.org/10.1101/gad.285395.116
- Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci 2005; 30(5):256-263; PMID:15896744; https://doi.org/10.1016/j.tibs.2005.03.009
- Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, de Murcia G, Evans R, Chambon P, Reinberg D. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell 2005; 18(1):83-96; PMID:15808511; https://doi.org/10.1016/j.molcel.2005.02.034
- Hsieh TH, Weiner A, Lajoie B, Dekker J, Friedman N, Rando OJ. Mapping Nucleosome Resolution Chromosome Folding in Yeast by Micro-C. Cell 2015; 162(1):108-119; PMID:26119342; https://doi.org/10.1016/j.cell.2015.05.048
- Schneider M, Hellerschmied D, Schubert T, Amlacher S, Vinayachandran V, Reja R, Pugh BF, Clausen T, Köhler A. The Nuclear Pore-Associated TREX-2 complex employs mediator to regulate gene expression. Cell 2015; 162(5):1016-1028; PMID:26317468; https://doi.org/10.1016/j.cell.2015.07.059
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 2013; 494(7438):497-501; PMID:23417068; https://doi.org/10.1038/nature11884
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013; 498(7455):516-520; PMID:23728302; https://doi.org/10.1038/nature12210
- Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature 2000; 408(6809):225-229; PMID:11089979; https://doi.org/10.1038/35041603
- Tantale K, Mueller F, Kozulic-Pirher A, Lesne A, Victor JM, Robert MC, Capozi S, Chouaib R, Bäcker V, Mateos-Langerak J et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat Commun 2016; 7:12248; PMID:27461529; https://doi.org/10.1038/ncomms12248
- Peng J, Zhou JQ. The tail-module of yeast Mediator complex is required for telomere heterochromatin maintenance. Nucleic Acids Res 2012; 40(2):581-593; PMID:21930512; https://doi.org/10.1093/nar/gkr757
- Zhu X, Liu B, Carlsten JO, Beve J, Nyström T, Myers LC, Gustafsson CM. Mediator influences telomeric silencing and cellular life span. Mol Cell Biol 2011; 31(12):2413-2421; PMID:21482672; https://doi.org/10.1128/MCB.05242-11

