Abstract
Pongamia pinnata is a species native to Indonesia and a potential bioenergy species to be developed on marginal land in Indonesia. This study examined variability in the pods, seeds, and oil content of 44 candidate plus trees (CPT) collected from a natural distribution of pongams on three islands and pongam domestication plots. The measurements of the phenotypic properties of the pods and seeds were recorded (length, width, thickness, and weight), as well as the oil content of the seeds. This study showed that Pongamia pinnata in Indonesia has variable pod and seed traits. However, there was no significant correlation between the pods/seeds trait with the oil content, indicating that the size of the seed does not determine oil content. The narrow-sense heritability of seed weight and oil content were high. The cluster of 44 CPT referred to oil content and 17 CPT can be promoted for breeding program. This study will help better identify for tree selection in a pongam breeding program.
1. Introduction
Fatty oils are a good energy source as they can be converted into biofuels in the renewable petrochemical industry. Tree species that will be developed for biofuels in Indonesia are determined based on the following criteria: (1) produces fatty oils, (2) value-added products (and adds economic value), and (3) grown on land that is not suitable for oil palm cultivation (Wargadalam et al., Citation2015). Other criteria for developing potential energy crops for the marginal lands of Indonesia include oil productivity, high energy wood, N2 fixing ability, non-oil added value, fast-growing, and capability of growing in salty/seawater, and only Pongamia pinnata meets these criteria (Lamria, Citation2019).
Pongamia pinnata L. Pierre, also known as pongam, or malapari, is a native species in Indonesia. It inhabits the coasts of Sumatera, Java, Madura, Bali, Nusa Tenggara, and Kalimantan (Leksono et al., Citation2021). In India, pongam are found from the coast to altitudes above 1,000 m (Sunil et al., Citation2012). These species bind nitrogen from the air through symbiosis with Rhizobium (Samuel et al., Citation2013). This allows pongam to be grown on marginal soil and for land rehabilitation purposes because of its ability to improve soil fertility (Edrisi and Abhilash, Citation2016; Ramachandran and Radhapriya, Citation2016; Borchard Citation2018; Yu et al., Citation2021, Citation2022).
Pongam is mainly utilized as biofuel and medicine. The oil is obtained from the seed by mechanical pressing or n-hexane extraction (Kumar and Kaushik, Citation2015; Sharma et al., Citation2016). Pongam seeds contain oil with 23%–26% fat content (Aminah et al., Citation2017). Oleic acid is the main fatty acid in pongam oil (Mukta et al., Citation2009; Rahangdale et al., Citation2014; Sharma et al., Citation2016; Aminah et al., Citation2017), and can be processed into biodiesel (Rengasamy et al., Citation2014). The oilseed residual waste is processed into ethanol through fermentation (Doshi and Srivastava, Citation2013; Radhakumari et al., Citation2017). All parts of the pongam plant are used by traditional communities for medicine (Chopade et al., Citation2008; Yadav et al., Citation2011). The active substances in pongam, such as pongamol and caranjin, are used as medicines and pesticides (Chopade et al., Citation2008; Sharma et al., Citation2020).
Pongam plantations are required for commercial-scale operations; however there are still some issues to be concerned in term of developing pongam program. Selecting Pongamia trees based on the selection of characteristics that close linkage with kernel yield. A pongam breeding program must be implemented to produce pongam plus/elite trees because the productivity of pongam seeds and oil is low but highly variable (Dalemans et al., Citation2022). The impact of abiotic factors such as heat (Cortes et al., Citation2020), salinity (Arpiwi et al., Citation2012) and biotic factors i.e pathogens, weeds, insect and pest (Escudero et al., Citation2021) should be taken into account. Pongam breeding must be supported by genetic material/genetic diversity from proven sources to intensify selection (Divakara, et al., Citation2011). Studying pod and seed morphological characters is often helpful when assessing genetic diversity (Mukta et al., Citation2009; Divakara et al., Citation2010; Sunil et al., Citation2012; Rao et al., Citation2008; Jaisankar et al., Citation2014; Patil and Naik, Citation2016).
In this study, we analyzed morphological data of pod and seed and its oil content to answer the following questions: (1). What is the diversity of pongam based on pod and seed traits, (2) what is the correlation between pod and seed phenotypic characters to oil content, (3) which trait can be used for selection and its heritability, and (4) which CPT should be promoted for breeding program.
2. Materials and methods
2.1. Location
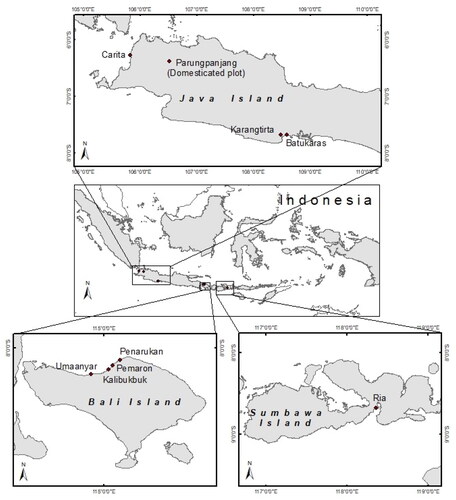
Candidate plus trees (CPT) were collected from their natural distribution and domesticated plots. Pongam flowering and fruiting occur asynchronously (Arpiwi et al., Citation2014); CPT was selected from their natural distribution during a one-time exploration. CPT was sampled from the domesticated plots during 1 year of observations. A total of 44 CPT were collected ().
Table 1. Origin and number of candidate tree plus
2.2. Pod and seed measurement
A total of 100 pods were collected from each CPT. Pod divided into four groups as replication for measurement of pod and seed traits. The pod and seed phenotypic characters were recorded for length, width, thickness, and weight. The oil content of the seeds was included as a seed trait.
2.3. Oil seed extraction
The seeds were air-dried and then oven-dried to reach 6%–8% moisture content. The oil was extracted using a mechanical press at a press temperature of 200 °C. The oil content value was the amount of oil (g) divided by the weight of the seeds (g).
2.4. Data analysis
Pod and seed traits were evaluated by analysis of variance. Tukey’s test was used as a posthoc test when the F test was significant. Pearson’s correlation coefficient analysis (P ≤ 0.05) was performed to analyze the linear correlations between the traits. Narrow sense heritability (h2) of pod and seed traits was compute using the additive (σa2) and residual (σe2) variances using equation h2 = σa2/(σa2 + σe2) (Fernández-Paz et al., Citation2021, Galeano et al., Citation2023) Cluster analysis were used to explain the pattern of variation among the CPT. Minitab 20 was used for the data analysis and a cluster is constructed with 500 bootstraps using R version 4.3.2 software.
3. Results
3.1. Variability in the pod and seed traits
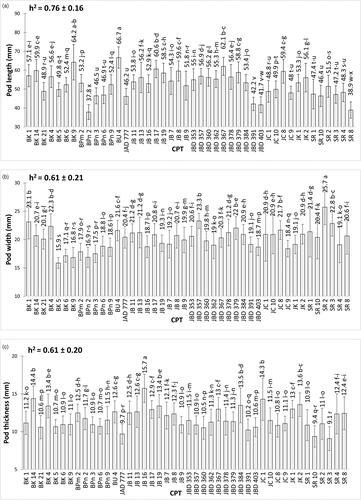
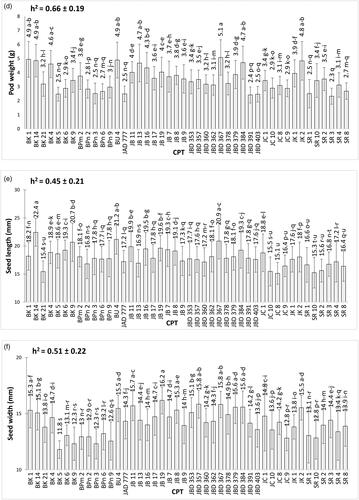
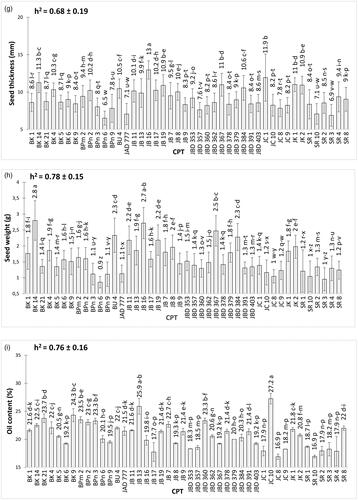
The pod and seed traits (pod length, pod width, pod thickness, pod weight, seed length, seed width, seed thickness, and seed weight) and total oil content were significantly different among the 44 CPT (). Of the nine characters observed, the highest character values were detected in seven CPT; but the lowest values were possessed by five CPT.
Figure 2. Mean values (± SE) of pod and seed traits on 44 candidate plus trees: (a) pod length, (b) seed length, (c) pod width, (d) seed width, (e) pod thickness, (f) seed thickness, (g) pod weight, (h) seed weight, (i) oil content. Different letters on bar indicate a significant difference using Tukey test at p < 0.05. Narrow-sense heritability (h2) is also included in the figures.
Variability studies revealed that CPT JB 16 had the maximum recorded values for pod thickness (15.73 mm) and pod width (12.96 mm), whereas BK 14 had the maximum values for seed length (22.44 mm) and seed weight (2.8 g). The longest pod was detected in BU 4 (66.74 mm). The widest pod was detected in SR2 (25.73 mm). JB 13 had the heaviest pod (5.08 g). JB 19 had the widest seeds (16.19 mm), and JC 10 had the maximum oil content (27.24%). However, the lowest pod and seed trait values were recorded for genotypes BK 5, BPn 2, BPn 6, JC 8, and SR 3, as shown in .
3.2. Correlations between the pod and seed traits
The relationship between the two attributes was assessed by Pearson’s correlation analysis (). The results showed that the direction of the correlation between traits was generally positive with a robust 0.5 < r < 0.75) to powerful (r > 0.75) correlation. However, no significant correlations were found between the pod/seed traits and oil content, as shown in .
Table 2. Pearson’s correlation coefficients between variables
3.3. Narrow-sense heritability of pod and seed traits
According to Comstock (Citation1949) heritability was classified into low (0%–30%), moderate (31%–60%), and high (> 60%) classes. All factors investigated have high levels of narrow sense heritability (h2), with the exception of the seed length and seed width, which are at a moderate level ().
3.4. Cluster analysis
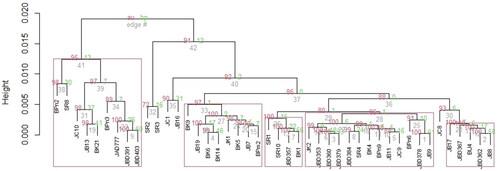
shows the dendrogram along with approximately unbiased (AU) p-values and probability bootstrap (BP) values. There are 5 groups that are interpreted as significant groups with a significance level of 0.05. There is no single group that pooled CPTs with the same geographic area. The clustering of 44 CPT revealed that several trees with the same origin were clustered in one group, but there were always other CPT grouped outside the group. Five CPT namely SR2, SR3, JC1, JB16, and JC8 located outside the group.
Figure 3. Clustering of 44 candidate plus trees (CPT) according to the pod and seed traitsRemarks: The values on the edges of the clustering represent p-values (%). The redcolored values show the AU (approximately unbiassed) p-values while the green-colored ones mean the BP (bootstrapt probability) values. Four red-colored rectangles in a bootstrap probability clustering represent clusters with the AU larger than 95%, which means that the results show rather strong correlation by the data.
4. Discussion
Analysis of variance for pod and seed traits revealed significant variation among the CPTs for all characters (P ˂ 0.05). Variations in the pods and seeds have also been reported by previous studies (Mukta et al., Citation2009; Divakara et al., Citation2010; Sunil et al., Citation2012; Rao et al., Citation2008; Jaisankar et al., Citation2014; Patil and Naik, Citation2016).
This study showed high variability in the pod and seed traits, particularly pod/seed weights. Previous studies also reported high variability in pod/seed weights (Sunil et al., Citation2012; Jaisankar et al., Citation2014) and other species, such as jatropha (Kaushik et al., Citation2007b). The correlation matrix revealed no correlation between seed properties and oil content, as reported previously (Sunil et al., Citation2012; Arpiwi et al., Citation2012; Sharma et al., Citation2016). The absence of a significant correlation between seed and oil content means that seed size does not determine oil content. However, larger seeds are preferred to smaller seeds (Kaushik et al., Citation2007a; Kesari et al., Citation2008). The preference for giant seeds was supported by Patil and Naik (Citation2016) who reported a positive correlation between pod/seed traits and oil content. Kaushik et al. (Citation2007b) stated that seed weight is positively and significantly correlated with oil content for other tree borne oil seed like jatropha.
Without a correlation between oil content and seed weight, the criteria for selecting a promising material for breeding programs could be considering both traits. Initial selection could be made using high seed weight and oil content. Trees with heavy seeds and high oil yields could be used as superior clones, which would be further prioritized for cultivation. These trait supported by narrow-sense of heritability that show hight heritablity for seed weight (h2 = 0.78 ± 0.15) and oil content (h2 = 0.76 ± 0.16) ().
Based on clustering, we found out that the 5 group refers by oil content. CPT with high oil content is distributed in group 1 (BPn2, SR8, JC10, JB13, Bk21, BPn3, JAD777, JBD391, JBD403) with an oil content range of 20 ̶ 25.87% and group 2 (BK9, JB19, BK6, BK14, JK1, BK5, JB7, BPm2) with an oil content between 19.24─24.31%. On the contrary, CPT with high seed weight was spread into 5 groups. Based on this result, the CPT on these two groups can be promoted as plus tree and selection in the breeding program should be focused on oil content.
This study also showed that geographic diversity did not necessarily represent genetic diversity. This result aligns with the P. pinnata grouping pattern in India (Kaushik et al., Citation2007a; Jaisankar et al., Citation2014) or jatropha in India (Kaushik et al., Citation2007b; Rao et al., Citation2008).
This research is concerned with selecting Pongamia trees that have a higher level of productivity than the average, based on phenotypic selection which is closely related to kernel yield. Sharma et al. (Citation2017) stated that pongam are mostly crossing species with low levels of selfing. This implies that its diversity is very high and opens up many possibilities for genetic improvement. Integrated superior seed traits from one exotic ancestry into a more elite background traits can be obtained by crossing, whether using a backcross schemes and and moder genomic-assisted backcrossing (Cortes et al. Citation2020).
5. Conclusion
This study showed that P. pinnata in Indonesia has variable pod and seed traits. The purpose of breeding pongam is to obtain trees with high yield and oil content, and no significant correlations were found between the pod/seed traits and oil content. The narrow-sense heritability of seed weight and oil content were high. The cluster of 44 CPT referred to oil content and 17 CPT can be promoted for breeding program. This study will help better identify for tree selection in a pongam breeding program.
Author contributions
D, HSN, DDNC, AA: planned the experiments; D, HSN, DDNC, AA, DS, NS, ES, DD, AHL, KAH collected samples and data measurements; D, DDNC, HSN performed data analysis; D, HSN, DDNC wrote the original draft preparation; D, HSN, DDNC, AA, DS, NS, ES, DD, AHL, KAH: interpreted and discussed the results; DS, AHL, KAH: validation and editing.
Acknowledgment and/or disclaimers
We thank The Head of Office for Standard Implementation of Environment and Forestry Instruments of Bogor (BPSI Bogor), Ministry of Environment and Forestry (MoEF), for the permission to access the domestication plot in the Parung Panjang Forest Research. This study was supported by the National Research and Development Agency (BRIN) for Bali and Sumbawa’s exploration (PEE Grand no 22119892994) and Rumah Program Pengungkapan dan Pemanfaatan Biodiversitas Nusantara (2023). Java’s exploration was supported by PT Sahabat Nusantara Teknologi Inovasi (PT SANTI). The authors declare no conflict of interest. All authors have read and agreed to the published version of the manuscript.
References
- Aminah A, Supriyanto, Siregar IZ, Suryani A. 2017. Kandungan minyak malapari (Pongamia pinnata (L.) Pierre) dari Pulau Jawa sebagai sumber bahan baku biodiesel. J Penelitian Hasil Hutan. 35(4): 255–262. doi: 10.20886/jphh.2017.35.4.255-262.
- Arpiwi NL, Yan G, Barbour EL, Plummer JA. 2012. Genetic diversity, seed traits and salinity tolerance of Millettia pinnata (L.) Panigrahi, a biodiesel tree. Genet Resour Crop Evol 60, 677–692 (2013). doi: 10.1007/s10722-012-9866-y.
- Arpiwi NL, Yan G, Barbour EL, Plummer JA. 2014. Phenology, pollination and seed production of Millettia pinnata in Kununurra, Northern Western Australia. J Biologi. 18:19–23.
- Borchard N, Bulusu M, Hartwig AM, Ulrich M, Lee S, Baral H. 2018. Screening potential bioenergy production of tree species in degraded and marginal land in the tropics. Forests. 9(10), 594. doi: 10.3390/f9100594.
- Chopade VV, Tankar AN, Pande VV, Tekade AR, Gowekar N, Bhandari S, Khandake SN. 2008. Pongamia pinnata: Phytochemical constituents, traditional uses and pharmacological properties: A review. Itl J of Green Pharmacy. 2(2):72–75. doi: 10.4103/0973-8258.41173.
- Comstock RE, Robinson HF, Harvey PH. 1949. A breeding procedure designed to make maximum use of both general and specific combining ability. Agronomy J. 41(8):360–367. doi: 10.2134/agronj1949.00021962004100080006x.
- Cortes AJ, Hernandez FL, Rodrigues DO. 2020. Prodicting thermal adaptation by looking into population genomic past. Frontiers in Genetics. 11:564515. doi: 10.3389/fgene.2020.564515.
- Dalemans F, Fremout T, Gowda B, Meerbeek KV, Muys B. 2022. Tempering expectations on a novel biofuel tree: Seed and oil yield assessment of pongamia (Millettia pinnata) shows low productivity and high variability. Industrial Crops and Products. 178:114384. doi: 10.1016/j.indcrop.2021.114384.
- Divakara BN, Alur AS, Tripati S. 2010. Genetic variability and relationship of pod and seed traits in Pongamia Pinnata (L.) Pierre., a potential agroforestry tree. Itl J of Plant Production. 4(2):129–141.
- Divakara BN, Upadhyaya HD, Krishnamurthy R. 2011. Identification and evaluation of diverse genotypes in Pongamia pinnata (L.) Pierre for genetic improvement in seed traits. J of Biodiversity and Ecological Sci. 1(3):179–190.
- Doshi P, Srivasta G. 2013. Sustainable approach to produce bioethanol from Karanja (Pongamia pinnata) oilseed residue. Turkish J of Agriculture and Forestry. 37(6):781–788. doi: 10.3906/tar-1207-18.
- Edrisi SA, Abhilash PC. 2016. Exploring marginal and degraded lands for biomass and bioenergy production: An Indian scenario. Renewable and Sustainable Energy Reviews. 54:1537–1551. doi: 10.1016/j.rser.2015.10.050.
- Escudero MG, Osorio AN, Cortes AJ. 2021. Integrative pre-breeding for biotic resistance in forest trees. Plants. 10(10). doi: 10.3390/plants10102022.
- Fernández-Paz J, Cortés AJ, Hernández-Varela CA, Mejía-de-Tafur MS, Rodriguez-Medina C, Baligar VC. 2021. Rootstock-mediated genetic variance in cadmium uptake by juvenile cacao (Theobroma cacao L.) genotypes, and its effect on growth and physiology. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.777842.
- Galeano E, Bockstette S, Thomas BR. 2023. An economic analysis and seed yield assessment of alternative breeding strategies in a white spruce tree improvement program. The Forestry Cronicle. 99(1):52–66. doi: 10.5558/tfc2023-001.
- Jaisankar I, Sankaran M, Singh DR, Damodaran V. 2014. Genetic variability and divergence studies in pod and seed traits of Pongamia pinnata (L.) Pierre., accessions in Bay Islands. J of Forestry Research. 25(2):351–358. doi: 10.1007/s11676-013-0422-1.
- Kaushik N, Kumar S, Kumar K, Beniwal RS, Kaushik N, Roy S. 2007a. Genetic variability and association studies in pod and seed traits in Pongamia pinnata (L.) Pierre in Haryana India. Genetic Resources and Crop Evolution. 54:1827–1832. doi: 10.1007/s10722-006-9204-3.
- Kaushik N, Kumar K, Kumar S, Kaushik N, Roy S. 2007b. Genetic variability and divergence studies in seed traits and oil content of Jatropha (Jatropha curcas L.) accessions. Biomass and Bioenergy. 31(7):497–502. doi: 10.1016/j.biombioe.2007.01.021.
- Kesari V, Krishnamachari A, Rangan L. 2008. Systematic characterization and seed oil analysis in candidate plus trees of biodiesel plant, Pongamia pinnata. Annals of Applied Biology. 152(3):397–40 doi: 10.1111/j.1744-7348.2008.00231.x.
- Kumar R, Kaushik N. 2015. Variability for seed oil content and seedling traits in Pongamia pinnata (L.) Pierre. J of Applied and Natural Science 7(2):1036–1041. doi: 10.31018/jans.v7i2.727.
- Lamria M. 2019. Increasing liquid fuel self-sufficiency in Indonesia through utilization of marginal land and appropriate technology for biofuel production. Doctor Thesis, Massey University, New Zealand.
- Leksono B, Rahman SA, Larjavaara M, Purbaya DA, Arpiwi NL, Samsudin YB, Artati Y, Windyarini E, Sudrajat DJ, Aminah A, Maulana AM, Bhatta KP, Kwon J, Baral H. 2021. Pongamia: A possible option for degraded land restoration and bioenergy production in Indonesia. Forests. 12(11):1468. doi: 10.3390/f12111468.
- Mukta N, Murthy IYLN, Sripal P. 2009. Variability assessment in Pongamia pinnata (L.) Pierre germplasm for biodiesel traits. Industrial Crops and Products. 29(2-3):536–540. doi: 10.1016/j.indcrop.2008.10.002.
- Patil VK, Naik GR. 2016. Variability in pod and seed traits of Pongamia pinnata Pierre ecotypes in North Karnataka, India. J of Forestry Research 27:557–567. doi: 10.1007/s11676-015-0191-0.
- Radhakumari M, Taha M, Shahsavari E, Bhargava SK, Satyavathi B, Ball AS. 2017. Pongamia pinnata seed residue: A low cost inedible resource for on-site/in-house lignocellulases and sustainable ethanol production. Renewable Energy. 103:682–687. doi: 10.1016/j.renene.2016.10.082.
- Rahangdale CP, Koshta LD, Patle NK. 2014. Provenance variation for oil content and fatty acid composition in seed of Pongamia pinnata (L.) Pierre. Itl J of Project Management. 4(12):1–9.
- Ramachandran A, Radhapriya P. 2016. Restoration of degraded soil in the Nanmangalam Reserve Forest with native tree species: Effect of indigenous plant growth-promoting bacteria. The Scientific World Journal. 2016:5465841. doi: 10.1155/2016/5465841.
- Rao GR, Korwar GR, Shanker AK, Ramakrishna YS. 2008. Genetic associations, variability and diversity in seed characters, growth, reproductive phenology and yield in Jatropha curcas (L.) accessions. Trees. 22:697–709. doi: 10.1007/s00468-008-0229-4.
- Rengasamy M, Anbalagan K, Mohanraj S, Pugalenthi V. 2014. Biodiesel production from Pongamia pinnata oil using synthesized iron nanocatalyst. International Journal of ChemTech Research. 6(10):4511–4516.
- Sunil N, Kumar V, Sivaraj N, Abraham B, Panwar NS, Varaprasad KS. 2012. Identification of areas of diversity and distribution of Pongamia based on altitude and seed traits. Indian J of Agricultural Sci. 82(6):489–93. doi: 10.56093/ijas.v82i6.18871.
- Samuel S, Scott PT, Gresshoff PM. 2013. Nodulation in the legume biofuel feedstock tree Pongamia pinnata. Agricultural Research. 2(3):207–214. doi: 10.1007/s40003-013-0074-6.
- Sharma SS, Islam MA, Malik AA, Kumar K, Negi MS, Tripathi SB. 2016. Seed traits, fatty acid profile and genetic diversity assessment in Pongamia pinnata (L.) Pierre germplasm. Physiology and Molecular Biology of Plants. 22:193–205. doi: 10.1007/s12298-016-0356-0.
- Sharma, SS, Islam, MA, Negi MS,Tripathi SB. 2017. Estimation of Outcrossing Rates in Biodiesel Species Pongamia pinnata Based on AFLP and Microsatellite Markers. Natl. Acad. Sci. Lett. 40: 105–108. doi: 10.1007/s40009-016-0533-2.
- Sharma A, Kaushik N, Rathore H. 2020. Karanja (Milletia pinnata (L.) Panigrahi): a tropical tree with varied applications. Phytochemistry Reviews. 19:643–658. doi: 10.1007/s11101-020-09670-z.
- Wargadalam VJ, Sasti H, Trylucky DN, Hambalilamria E, Thahar A, Nisya FN. 2015. R&D roadmap of biofuel: towards energy self-sufficiency. Ministry of Energy and Mineral Resources.
- Yadav RD, Jain SK, Alok S, Prajapati S, Verma A. 2011. Pongamia pinnata: An Overveiw. Itl J of Pharmaceutical Sciences and Research. 2(3):494–500. doi: 10.13040/IJPSR.0975-8232.2(3).494-00.
- Yu X, Shen T, Kang X, Cui Y, Chen Q, Shoaib M, Liu H, Zhang F, Hussain S, Xiang Q, Zhao K, Gu Y, Ma M, Li S, Zou L, Liang Y. 2021. Long-term phytoremediation using the symbiotic Pongamia pinnata reshaped soil micro-ecological environment. Sci of The Total Environment. 774:145112. doi: 10.1016/j.scitotenv.2021.145112.
- Yu X, Shoaib M, Cheng X, Cui Y, Hussain S, Yan J, Zhou J, Chen Q, Gu Y, Zou L, Zhang X, Hao S, Zhao K, Ma M, Xiang Q, Li S, Zou T. 2022. Role of rhizobia in promoting non-enzymatic antioxidants to mitigate nitrogen-deficiency and nickel stresses in Pongamia pinnata. Ecotoxicology and Environmental Safety. 241:113789. doi: 10.1016/j.ecoenv.2022.113789.