Abstract
Objective
Variants in the superoxide dismutase (SOD1) gene are among the most common genetic causes of amyotrophic lateral sclerosis. Reflecting the wide spectrum of putatively deleterious variants that have been reported to date, it has become clear that SOD1-linked ALS presents a highly variable age at symptom onset and disease duration.
Methods
Here we describe an open access web tool for comparative phenotype analysis in ALS: https://sod1-als-browser.rosalind.kcl.ac.uk/. The tool contains a built-in dataset of clinical information from 1383 people with ALS harboring a SOD1 variant resulting in one of 162 unique amino acid sequence alterations and from a non-SOD1 comparator ALS cohort of 13,469 individuals. We present two examples of analyses possible with this tool, testing how the ALS phenotype relates to SOD1 variants that alter amino acid residue hydrophobicity and to distinct variants at the 94th residue of SOD1, where six are sampled.
Results and conclusions
The tool provides immediate access to the datasets and enables bespoke analysis of phenotypic trends associated with different protein variants, including the option for users to upload their own datasets for integration with the server data. The tool can be used to study SOD1-ALS and provides an analytical framework to study the differences between other user-uploaded ALS groups and our large reference database of SOD1 and non-SOD1 ALS. The tool is designed to be useful for clinicians and researchers, including those without programming expertise, and is highly flexible in the analyses that can be conducted.
Background
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by dysfunction and death of motor neurons leading to progressive muscle weakness and paralysis (Citation1). Its clinical presentation can vary greatly. For example, although people most frequently develop the first symptoms between 55 and 65 years of age, the disease can onset across all stages of adulthood. Similarly, time from symptom onset until death is a median of 3 years for ALS but some people die within a year of onset, and 5–10% of people survive for more than 10 years (Citation2–4).
A plethora of genetic factors can affect the risk of ALS or its phenotype, and mutations in specific genes can lead to distinct clinical outcomes. For example, a hexanucleotide repeat expansion in the C9orf72 gene is the most common known cause of ALS, and carriers of this mutation typically develop ALS earlier and with faster progressing symptoms than sporadic ALS patients (Citation5–8). Furthermore, different mutations within the same gene can also lead to distinct forms of the disease. For example, over 180 variants in the superoxide dismutase (SOD1) gene (Citation9–11) have been found in ALS patients. As well as affecting ALS risk, some of these variants have distinct effects on clinical features such as the age of onset of motor symptoms and disease duration. For example, p.A5V and p.H44R have a marked effect on disease duration while p.G38R is associated with an early onset (Citation10,Citation12–14). Being able to characterize how genetic variants affect the clinical phenotype is essential for optimal development and design of healthcare, treatments, and trial stratification. However, the multitude of genetic factors involved in ALS and their rarity are great challenges for their individual study. To address these limitations, focussing on SOD1 given the recent gene therapy trials (Citation15), we recently collated data from the literature and specialized ALS centers globally on approximately 15,000 people with ALS, over 1000 of whom harbored a variant in the SOD1 gene (Citation10).
In this paper, we describe a web tool (https://sod1-als-browser.rosalind.kcl.ac.uk/) with upload facilities to allow people to perform comparative and bespoke phenotype analyses using data from a database of almost 15,000 people with ALS without the need for informatics proficiency. The tool currently allows users to define and select subgroups of patients with or without variants in SOD1, to stratify by individual or groups of SOD1 variants, and to upload data to combine with our database in the analysis. To show the potential of this tool and how to use it, we present two example case studies that leverage the data from our recent publication which is accessible to all users. The first example builds upon research suggesting that variants affecting protein hydrophobicity promote aggregation of mutant SOD116 and tests how alterations in amino acid hydrophobicity affect the ALS phenotype. The second example focuses specifically on variation at the 94th amino acid residue of SOD1, which is a site containing multiple reported variants, testing how the phenotype differs for each variant sampled.
Materials and methods
Dataset
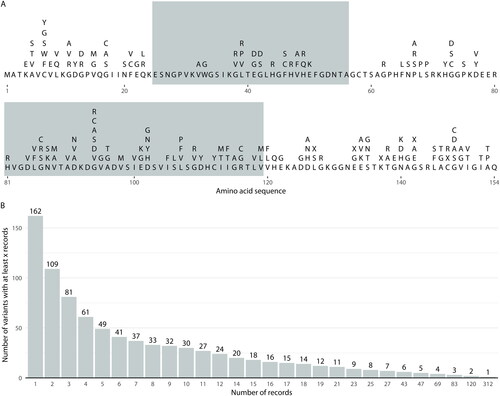
The tool enables access to a dataset of 14,852 people with ALS, 1383 of whom harbor a potentially deleterious non-synonymous SOD1 gene variant (N without SOD1 variant = 13,469). A total of 162 unique amino acid variants (canonical SOD1 sequence IDs: ENSEMBL = ENST00000270142.11, UniProt = P00441) are represented within these data (see ; Table S1). The dataset is further described within our previous publication (Citation10) and a summary of the disease characteristics associated with the 49 variants harbored by at least 5 people is provided on the site.
Results
Amino acid hydrophobicity analysis
In case study 1, we examined how the ALS phenotype varied by changes in amino acid hydrophobicity. Across all amino acid substitutions sampled: 42.86% were variants that remained in the same hydrophobicity category as wild type SOD1; 42.11% were variants with a hydrophilic or hydrophobic amino acid in the wild type and an intermediate amino acid in the mutant protein; 12.59% were variants with an intermediate amino acid becoming hydrophilic or hydrophobic; and 2.44% were variants with substitutions from hydrophilic to hydrophobic amino acids or vice versa (see ).
Table 1. Data summary for case studies. For disease duration analysis, restricted mean and median estimates are obtained from the survival curve and the standard error (SE) and 95% confidence interval (CI) indicates certainty in this estimate; the distribution of disease duration in non-censored individuals is reported directly. In age of onset analysis no person is censored, therefore estimates correspond with the raw descriptive statistics. *Minimum and maximum shown because disease duration was only available for two people. SD = standard deviation.
Figure 1 Variant characteristics for the native dataset. Panel A: The canonical SOD1 amino acid sequence (bold) and variants recorded at each residue, denoted using IUPAC amino acid nomenclature (Citation16), where “X” indicates protein truncating variants. Alternating background shading indicates residues encoded from different exons of the SOD1 gene. Panel B: The number of variants with at least a certain number of records available across different thresholds.
Functionality
Survival analysis methods can be performed for (1) age at symptom onset, and (2) disease duration from symptom onset (with a corresponding censor variable indicating survival status). Kaplan-Meier and Cox proportional-hazards (CPH) approaches are both implemented and relevant descriptive statistics for the analyzed sample are given by strata. Differences between strata in univariate analyses are examined using the log-rank test; global and pairwise log-rank tests are performed when more than two strata are defined. Analyses using CPH models are performed whenever two or more strata are defined or when a single stratum is specified and the user selects at least one of several covariates which can be included in the regression model. Available covariates are clinical diagnosis, family disease history, sex, age of onset, site of onset, and sample source (continent of origin). Users can pick which covariates are included in the analysis depending on requirements and associations between selected survival analysis strata and available covariates can be tested.
Various analysis options are provided. The user can model survival for any number of individual SOD1 variants (including a “no variant” option) and variants can be collapsed into groups of interest (including an “any other SOD1 variant” option). We include three pre-defined options for grouping variants: by functional location (Citation10) in the protein (across the dimer interface, electrostatic loop, zinc loop, and other) or according to the gene exon from which variants are transcribed. The final pre-defined analysis compares people with any SOD1 variant versus the “no variant” group.
Users can further customize the analysis. They can filter by continent of origin and opt to stratify the analysis by sex, family history, site of onset, clinical diagnosis, and country or continent of origin. Time-dependent CPH analyses are also possible, allowing users to define timepoints at which the data are split. This functionality allows time-dependent coefficients to be modeled and enables analysis constrained to a certain timeframe (e.g. only the first 12 months from disease onset).
We allow users to upload supplemental data that is appended to the native sample, enriching the analysis possible within the tool. There are no restrictions regarding records that can be uploaded as supplemental data; users may provide data associated with SOD1 variants both present in and absent from the built-in dataset or provide data from other groups of patients (e.g. for variants from other genes). Formatting instructions for supplemental data are provided on the site.
The results of the user”s analyses are presented on the website, and we provide options to (1) download these within an HTML report and (2) download publication-ready versions of the figures produced, with customizable formatting.
Tool design
The tool is written in the R programming language (R version 4.2.3) and developed using the R packages (versions) shiny (1.7.4), shinyjs (2.1.0), shinycssloaders (1.0.0), tidyverse (2.0.0), rmarkdown (2.21), countrycode (1.4.0), kableExtra (1.3.4), and plotly (4.10.1) (Citation17–25). Survival analyses are performed and visualized using survival (version 3.5-5), and survminer (version 0.4.9) (Citation26,Citation27).
Examples of use
Here we present two examples of analyses possible within this tool. We examined differences in age of onset and disease duration between the strata of each example using Kaplan-Meier analyses and the log-rank test, and CPH models were applied with robust variance estimation as implemented by coxph to examine differences between strata before and after controlling for possible covariates. In the CPH models, we controlled for sex and age of onset when analyzing disease duration, and sex only when analyzing age of onset.
Case study 1 examined whether changes to amino acid hydrophobicity influenced the age of ALS onset or disease duration from onset until death. Amino acids were grouped into three hydrophobicity categories (Citation28): hydrophobic (Amino acid IUPAC code (Citation16): F, M, I, L, V), hydrophilic (D, E, H, K, R, N, Q), and intermediate (Y, W, P, G, A, S, T, C). Variants resulting in an amino acid substitution were then categorized based on the hydrophobicity group of the wild type and mutant amino acid; Table S1 presents the assignment of groups and data availability across variants. To specifically examine the consequence of changes in hydrophobicity, three sets of analyses were conducted, each respective to variants occurring in residues that are hydrophilic, intermediate, or hydrophobic in the wild-type protein. In each analysis, variants resulting in altered hydrophobicity were compared against variants where the mutant and wild type amino acids remained in the same hydrophobicity group. The p.A5V variant was excluded from these analyses since it is characterized by a particularly aggressive phenotype and accounted for the majority of records (n = 312) in the “intermediate to hydrophobic” category. A broader hydrophobicity analysis across all groups was also conducted.
Case study 2 examined trends associated with variation at the 94th amino acid residue of SOD1, coding for a glycine in the wild type protein. Six variants were present at this locus. We first analyzed differences in age of onset and disease duration associated with having any p.G94 variant vs any other SOD1 variant. Second, we compared p.G94 variants individually to non-p.G94 variants, aggregating across p.G94R, p.G94S, and p.G94V since they each contained fewer than 5 records. summarizes characteristics of the data from both case studies.
Age at symptom onset appeared roughly comparable across variants in all categories of the hydrophobicity analyses (see ; Table S2; ), with all groups having a mean age of onset between 46 and 51 years ().
Table 2 Inferential statistics for survival analyses across case studies. Bold values denote nominal p values <0.05. * controlling for sex in the age of onset analysis and for sex and age of onset in the disease duration analysis. #Hazard ratios greater than 1 indicate earlier age of onset/shorter disease duration in the non-reference group. †No p.G94S variants were available for the disease duration analysis. CPH = Cox proportional-hazards.
Figure 2 Kaplan-Meier survival curves for age of onset and disease duration analyses. Analysis shown: Panels A-B: trends associated with wild type and variant amino acid hydrophobicity; Panels C-D: Any SOD1 p.G94 variant versus non-p.G94 SOD1 variants (OtherVariant). Panels E-F: individual p.G94 variants versus non-p.G94 SOD1 variants. Panels A, C, and E are for age of onset analysis, and B, D, and F describe disease duration. Panels A and B display all hydrophobicity groups in a single figure for each analysis and confidence intervals are not displayed to maximize visual clarity; Figure S1 visualizes trends in age of onset and disease duration for these groups after stratifying across panels according to the hydrophobicity group of the wild type residue.
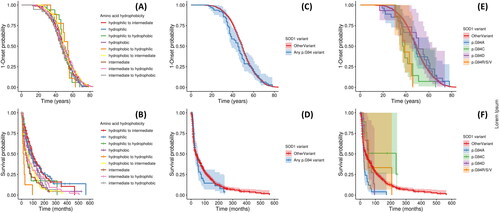
Disease duration analysis (see ; ) however, suggested that alterations in amino acid hydrophobicity may affect disease prognosis following onset. Analysis of variants at residues which are hydrophobic in wild type SOD1 indicated that disease duration was shorter in substitutions to hydrophilic amino acids (p value: log-rank test = 1.25 × 10−5; CPH model = 9.19 × 10−8) and that substitution into intermediate amino acids also tended toward shorter disease duration (p value: log-rank test = 0.0202; CPH model = 0.0503). The estimated median (95% CI) disease duration was 74 (50, 109) months for hydrophobic to hydrophobic substitutions, 45 (30, 72) for hydrophobic to intermediate, and 18 (10, NA) for hydrophobic to hydrophilic.
Among variants occurring in intermediate residues of wild-type SOD1, becoming either hydrophilic or hydrophobic was associated with longer disease duration. The estimated median disease duration (95% CI) for intermediate to intermediate amino acid substitutions was 38.4 (24, 53) months, under half that of intermediate to hydrophobic (84 (56, 168)) and shorter than intermediate to hydrophilic (45 (32, 84)) mutations.
Analysis of variants in hydrophilic residues of wild-type SOD1 did not identify clear differences in disease duration between substitutions that remained hydrophilic and those which became intermediate (p value: log-rank test = 0.619; CPH model = 0.209) or hydrophobic (p value: log-rank test = 0.296; CPH model = 0.289). The estimated median disease duration (95% CI) for hydrophilic to hydrophilic substitutions was 85 (66, 125) months, trending toward being shorter than in hydrophobic to intermediate (96 (84, 123)) and longer than in hydrophilic to hydrophobic (65 (39, NA)) substitutions.
Table S3 presents an additional CPH model comparing all hydrophobicity groups relative to substitutions in residues with intermediate to intermediate amino acid substitutions. The analysis indicated that disease duration was shortest in this and the hydrophobic to hydrophilic substitution groups.
p.G94 amino acid residue analysis
In case study 2, we examined trends associated with variation in the 94th SOD1 residue. p.G94A was the most frequent variant at this locus and 5 other variants occurred in the dataset (see ). This case study showed variant-specific trends in age of onset and disease duration, which were not discernable aggregating across p.G94 variants, when compared with non-p.G94 SOD1 variants (see ; ).
Age of onset was earlier than in the non-p.G94 SOD1 variant reference category only in the p.G94C (p value: log-rank test = 6.73 × 10−4; CPH model = 7.39 × 10−4) and p.G94R/S/V (p value: log-rank test = 5.66 × 10−4; CPH model = 9.34 × 10−8) groups; this difference appears considerable since the median age of onset for non-p.G94 SOD1 is over 10 years later than median onset in these two groups (see ; ).
The disease duration analysis indicated that only the p.G94A variant was associated with shorter time to death (p value: log-rank test = 5.95 × 10−3; CPH model = 3.00 × 10−5). Inspection of hazard ratios suggests that p.G94C trended toward longer disease duration compared to non-p.G94 variants even after controlling for age of onset and sex (p value: log-rank test = 0.0672; CPH model = 0.100). Although the median disease duration was longer for variants in the p.G94D and p.G94R/S/V variant groups, data were insufficient to test the association.
Discussion
We have developed a web-tool to facilitate bespoke investigations of the impact of SOD1 variants upon the ALS phenotype, using survival analysis approaches. We have provided two examples of this tool”s utility, examining differences in ALS age at symptom onset and disease duration according to (1) variants of varying impact upon residue hydrophobicity across SOD1 and (2) distinct variants at the 94th SOD1 residue.
This online facility has key benefits for research on the heterogenous ALS phenotype. First, it permits a user-friendly interface for performing survival analysis, with various options for customization in accordance with the user”s needs. Second, it provides access to a large in-built SOD1-ALS cohort and non-SOD1 comparator population, which can be further enriched if users provide their own supplementary data.
The hydrophobicity analysis suggested that substitution variants altering residue hydrophobicity from hydrophobic to intermediate or hydrophilic are associated with a shorter disease prognosis compared to variants in residues remaining hydrophobic across wild type and mutant SOD1. This aligns well with evidence that altered hydrophobicity promotes aggregation of the SOD1 protein (Citation29), and may reflect greater destabilization and misfolding of SOD1 when variants cause more extreme alterations in hydrophobicity (Citation30–32). Interestingly, variants of intermediate to intermediate amino acid substitutions were characterized by particularly short disease duration.
Hydrophobic to hydrophilic amino acid substitutions and vice versa were, notably, infrequent relative to other substitutions. Given that these would represent the most extreme hydrophobicity alterations, this could indicate a potential survivorship bias and that these substitutions may be sufficiently deleterious to be evolutionarily suppressed. This appears reasonable since SOD1 is highly conserved, with deficiency being linked to severe and early onset phenotypes (Citation33–35), and on the basis of variants in these hydrophobicity groups being entirely absent from the gnomAD v2.1.1 population database (Citation36) (see Table S4).
Analysis of the variants at p.G94 emphasized the extent to which individual SOD1 variants differentially influence the phenotype. Grouping together all p.G94 variants suggested that age of ALS onset and disease duration are comparable for people with variants at this residue and those with non-p.G94 SOD1 variants. Only by examining variants individually did we observe that p.G94A was associated with shorter, and p.G94C trended toward longer, disease duration than non-p.G94 SOD1-ALS. Likewise, p.G94C and the aggregation of p.G94R, p.G94S, and p.G94V were indicative of substantially earlier age of onset. These findings are consistent with the results of our previous analysis of SOD1-ALS, emphasizing distinction between trends in age of onset and disease duration across individual variants (Citation10). They highlight particularly the importance of making available resources to allow variant-level analyses of the ALS phenotype associated with variation in SOD1.
The tool is not without limitation. Most notable is that a number of the 162 SOD1 variants sampled are harbored by very few individuals and thus are not sufficient for individual variant analysis with the native dataset alone. However, this issue can be somewhat circumvented by aggregating rarer variants into a single analysis stratum, and by the possibility of increasing the dataset with user-supplied data.
Certain considerations apply when providing supplementary data to the tool. Firstly, CPH models may only include covariates that are available in the native dataset. Second, records from supplementary data may overlap with native dataset. To reduce this possibility, the tool will automatically flag any people among the supplementary dataset who may be a duplicate of a person in the built-in dataset, checking for matches by country of origin, SOD1 amino acid change (if the user indicates that one is present), age of onset, site of onset, sex, and disease duration (if not censored). Users can also consult the cohort description provided and contact ALSoD (https://alsod.ac.uk/ (Citation37);) with any concerns.
Overall, the open-access web-utility we provide (https://sod1-als-browser.rosalind.kcl.ac.uk/) has a potentially substantial benefit for ALS disease research and direct translational use for the design of patient stratification approaches, as well as being useful for mutation adjudication committees needing to make decisions on likely disease course with limited data. It permits an array of analysis options which can be readily implemented by users without any programming knowledge, and can be enriched by the provision of a supplementary dataset. Accordingly, this tool allows clinicians and researchers to circumvent many possible barriers they may otherwise face, for instance, regarding insufficient data availability or in preparing these data for analysis. The potential translational benefit of this tool is substantial, facilitating growth in understanding of the ALS phenotype which may aid the design and implementation of effective healthcare, treatments, and clinical trials.
Supplemental Table
Download MS Excel (27.6 KB)Acknowledgments
Samples used in this research were in part obtained from the UK National DNA Bank for MND Research, funded by the MND Association and the Wellcome Trust. Part of the samples were obtained from The Project MinE and STRENGTH consortia, and MND centers internationally. We would like to thank the members of these consortia and our clinical collaborators for their contribution: Philip van Damme, Philippe Corcia, Philippe Couratier, Patrick Vourc”h, Orla Hardiman, Russell McLaughin, Marc Gotkine, Vivian Drory, Nicola Ticozzi, Vincenzo Silani, Jan H Veldink, Leonard H van den Berg, Mamede de Carvalho, Jesus S Mora Pardina, Monica Povedano, Peter Andersen, Markus Weber, Nazli A Başak, Ammar Al-Chalabi, Christopher E Shaw, Pamela J Shaw, Karen E Morrison, John E Landers, Jonathan D Glass, Clifton L Dalgard, Nailah Siddique, Teepu Siddique, Kelly L Williams, Ian P Blair, Jennifer Jockel-Balsarotti, Lyndal Henden, Garth A Nicholson, Timothy Miller, Diane McKenna-Yasek, Robert H Brown, William Camu, Zorica Stevic, Lu Tang, Dong-sheng Fan. We thank people with MND and their families for their participation in this project. We acknowledge sample management undertaken by Biobanking Solutions funded by the Medical Research Council at the Center for Integrated Genomic Medical Research, University of Manchester. The authors acknowledge use of the research computing facility at King”s College London, Rosalind (https://rosalind.kcl.ac.uk), which is delivered in partnership with the National Institute for Health and Care Research (NIHR) Biomedical Research Centers at South London and Maudsley and Guy”s and St. Thomas” NHS Foundation Trusts, and part-funded by capital equipment grants from the Maudsley Charity (award 980) and Guy”s and St. Thomas” Charity (TR130505). The authors also acknowledge the use of the CREATE research computing facility at King”s College London (Citation38). We also acknowledge Health Data Research UK, which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (United Kingdom), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation and Wellcome Trust.
Declaration of interest
TS is employed by AstraZeneca. The other authors report no conflict of interest.
Data availability statement
All data associated with this manuscript can be accessed from the web-utility: https://sod1-als-browser.rosalind.kcl.ac.uk/.
Additional information
Funding
References
- Shatunov A, Al-Chalabi A. The genetic architecture of ALS. Neurobiol Dis. 2021; 147:105156.
- Chiò A, Logroscino G, Hardiman O, Swingler R, Mitchell D, Beghi E, et al. Prognostic factors in ALS: a critical review. Amyotroph Lateral Scler. 2009; 10:310–23.
- Juneja T, Pericak-Vance MA, Laing NG, Dave S, Siddique T. Prognosis in familial amyotrophic lateral sclerosis: progression and survival in patients with glu100gly and ala4val mutations in Cu, Zn superoxide dismutase. Neurology 1997; 48:55–7.
- Al-Chalabi A, Hardiman O. The epidemiology of ALS: a conspiracy of genes, environment and time. Nat Rev Neurol. 2013; 9:617–28.
- Byrne S, Elamin M, Bede P, Shatunov A, Walsh C, Corr B, et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol. 2012; 11:232–40.
- van Rheenen W, van der Spek RAA, Bakker MK, van Vugt JJFA, Hop PJ, Zwamborn RAJ, et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat Genet. 2021; 53:1636–48.
- Umoh ME, Fournier C, Li Y, Polak M, Shaw L, Landers JE, et al. Comparative analysis of C9orf72 and sporadic disease in an ALS clinic population. Neurology 2016; 87:1024–30.
- Murphy NA, Arthur KC, Tienari PJ, Houlden H, Chio A, Traynor BJ. Age-related penetrance of the C9orf72 repeat expansion. Sci Rep. 2017; 7:2116.
- Kalia M, Miotto M, Ness D, et al. Molecular dynamics analysis of superoxide dismutase 1 mutations suggests decoupling between mechanisms underlying ALS onset and progression. bioRxiv 2022. doi: https://doi.org/10.1101/2022.12.05.519128
- Opie-Martin S, Iacoangeli A, Topp SD, Abel O, Mayl K, Mehta PR, et al. The SOD1-mediated ALS phenotype shows a decoupling between age of symptom onset and disease duration. Nat Commun. 2022; 13:6901.
- Bunton-Stasyshyn RKA, Saccon RA, Fratta P, Fisher EMC. SOD1 function and its implications for amyotrophic lateral sclerosis pathology:new and renascent themes. Neuroscientist. 2015; 21:519–29.
- McCann EP, Williams KL, Fifita JA, Tarr IS, O”Connor J, Rowe DB, et al. The genotype–phenotype landscape of familial amyotrophic lateral sclerosis in Australia. Clin Genet. 2017; 92:259–66.
- Parton MJ, Broom W, Andersen PM, Al-Chalabi A, Nigel Leigh P, Powell JF, et al. D90A-SOD1 mediated amyotrophic lateral sclerosis: a single founder for all cases with evidence for a Cis-acting disease modifier in the recessive haplotype. Hum Mutat. 2002; 20:473.
- Bali T, Self W, Liu J, Siddique T, Wang LH, Bird TD, et al. Defining SOD1 ALS natural history to guide therapeutic clinical trial design. J Neurol Neurosurg Psychiatry. 2017; 88:99–105.
- Miller TM, Cudkowicz ME, Genge A, Shaw PJ, Sobue G, Bucelli RC, et al. Trial of antisense oligonucleotide tofersen for SOD1 ALS. N Engl J Med. 2022; 387:1099–110.
- IUPAC-IUB. Joint commission on biochemical nomenclature. Nomenclature and symbolism for amino acids and peptides: recommendations 1983. Eur J Biochem 1984; 138:9–37.
- Attali D. shinyjs: easily improve the user experience of your shiny apps in seconds. 2020. R package version 2.1.0. Available from: https://CRAN.R-project.org/package=shinyjs
- Chang W, Cheng J, Allaire J, et al. shiny: web application framework for R. 2022. R package version 1.7.4. Available from: https://CRAN.R-project.org/package=shiny
- R Core Team. R: A language and environment for statistical computing. 2021. Vienna, Austria: R Foundation for Statistical Computing,. Available from: https://www.R-project.org/
- Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, et al. Welcome to the Tidyverse. Joss. 2019; 4:1686.
- Allaire J, Xie Y, McPherson J, et al. rmarkdown: dynamic documents for R. 2022. R package version 2.21. Available from: https://rmarkdown.rstudio.com
- Arel-Bundock V, Enevoldsen N, Yetman C. Countrycode: an R package to convert country names and country codes. Joss. 2018; 3:848.
- Zhu H. kableExtra: construct complex Table with “kable” and Pipe Syntax. 2021. R package version 1.3.4. Available from: https://CRAN.R-project.org/package=kableExtra
- Sievert C. Interactive web-based data visualization with R, plotly, and shiny. Florida: CRC Press; 2020.
- Sali A, Attali D. shinycssloaders: Add Loading Animations to a “shiny” Output While It”s Recalculating. 2020. R package version 1.0.0. Available from: https://CRAN.R-project.org/package=shinycssloaders
- Kassambara A, Kosinski M, Biecek P. survminer: Drawing Survival Curves using “ggplot2”. 2021. 0.4.9. Available from: https://CRAN.R-project.org/package=survminer
- Therneau T. A Package for Survival Analysis in R. 2023. 3.5-5. Available from: https://CRAN.R-project.org/package=survival
- Sharer JD. Amino Acid Disorders. In: Aminoff MJ, Daroff RB, eds. Encyclopedia of the Neurological Sciences. Second Edition. Oxford: Academic Press; 2014. p. 136–47.
- Tompa DR, Kadhirvel S. Changes in hydrophobicity mainly promotes the aggregation tendency of ALS associated SOD1 mutants. Int J Biol Macromol. 2020; 145:904–13.
- Cordes MHJ, Sauer RT. Tolerance of a protein to multiple polar-to-hydrophobic surface substitutions. Protein Sci. 2008; 8:318–25.
- Gidalevitz T, Krupinski T, Garcia S, Morimoto RI. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLOS Genet. 2009; 5:e1000399.
- Dyson HJ, Wright PE, Scheraga HA. The role of hydrophobic interactions in initiation and propagation of protein folding. Proc Natl Acad Sci U S A. 2006; 103:13057–61.
- Park JH, Elpers C, Reunert J, McCormick ML, Mohr J, Biskup S, et al. SOD1 deficiency: a novel syndrome distinct from amyotrophic lateral sclerosis. Brain. 2019; 142:2230–7.
- Ezer S, Daana M, Park JH, Yanovsky-Dagan S, Nordström U, Basal A, et al. Infantile SOD1 deficiency syndrome caused by a homozygous SOD1 variant with absence of enzyme activity. Brain. 2022; 145:872–8.
- Farrimond L, Talbot K. A case of SOD1 deficiency: implications for clinical trials. Brain. 2022; 145:805–6.
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020; 581:434–43.
- Abel O, Powell JF, Andersen PM, Al-Chalabi A. ALSoD: A user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum Mutat. 2012; 33:1345–51.
- King”s College London. King”s computational research, engineering and technology environment (CREATE) 2022 [October 12 2022]. Available from: https://doi.org/10.18742/rnvf-m076
