?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background
Web search data have proven to bea valuable early indicator of COVID-19 outbreaks. However, the influence of co-morbid conditions with similar symptoms and the effect of media coverage on symptom-related searches are often overlooked, leading to potential inaccuracies in COVID-19 simulations.
Method
This study introduces a machine learning-based approach to estimate the magnitude of the impact of media coverage and comorbid conditions with similar symptoms on online symptom searches, based on two scenarios with quantile levels 10–90 and 25–75. An incremental batch learning RNN-LSTM model was then developed for the COVID-19 simulation in Australia and New Zealand, allowing the model to dynamically simulate different infection rates and transmissibility of SARS-CoV-2 variants.
Result
The COVID-19 infected person-directed symptom searches were found to account for only a small proportion of the total search volume (on average 33.68% in Australia vs. 36.89% in New Zealand) compared to searches influenced by media coverage and comorbid conditions (on average 44.88% in Australia vs. 50.94% in New Zealand). The proposed method, which incorporates estimated symptom component ratios into the RNN-LSTM embedding model, significantly improved COVID-19 simulation performance.
Conclusion
Media coverage and comorbid conditions with similar symptoms dominate the total number of online symptom searches, suggesting that direct use of online symptom search data in COVID-19 simulations may overestimate COVID-19 infections. Our approach provides new insights into the accurate estimation of COVID-19 infections using online symptom searches, thereby assisting governments in developing complementary methods for public health surveillance.
Introduction
The emergence of a novel coronavirus, severe acute respiratory syndrome coronavirus (SARS-CoV-2), in December 2019 has led to a global pandemic with devastating health and socio-economic impacts. According to the World Health Organisation (WHO), as of 1st March 2023, more than 758 million people have been infected, and 6.8 million have died from COVID-19, the disease caused by SARS-CoV-2 [Citation1,Citation2].
In response to this unprecedented public health crisis, researchers have leveraged online search data to provide real-time information and early detection of COVID-19 cases [Citation3–7]. Previous studies in the United States have found a significant correlation between online searches for COVID-19 symptoms and subsequent COVID-19 outbreaks [Citation3]. Another study has estimated a 15-day lag between online symptom search trends and COVID-19 case counts in nine countries [Citation8]. These findings suggest that online digital searches have the potential to improve the monitoring and forecasting of the COVID-19 epidemic and act as a valuable adjunct to existing COVID-19 surveillance systems [Citation6,Citation7]. Predominantly, the techniques in online search-based COVID-19 tracking analyses involves directly utilising an online search index within simulation models [Citation3,Citation4,Citation6–8]. However, this approach often fails to adequately account for various confounding factors [Citation5], which include (1) the presence of comorbid conditions that share similar symptoms with COVID-19 and (2) media coverage. Such omissions can potentially influence the interpretation of search data, leading to overestimating of COVID-19 case numbers. This highlights the need for a more nuanced approach to using online search data for disease tracking, where comorbidities and media influence are appropriately considered to improve the accuracy and reliability of results.
In particular, the presence of comorbid conditions with symptoms similar to those of COVID-19 can introduce significant noise and bias into data derived from online symptom searches and may lead to overestimating COVID-19 cases. Prior research shows a significant rise in online searches for symptoms like cough and sore throat during the influenza season [Citation9–11]. This study focuses on Australia and New Zealand, where flu season generally lasts from May to October [Citation12,Citation13]. Consequently, there’s an expected spike in online searches for symptoms common to the flu and other comorbid conditions during this period. This increase could lead to inflated COVID-19 infection estimates if online symptom search indexes are used directly in simulation models, thus affecting the accuracy of the results.
In addition, the impact of media coverage in shaping public search behaviours and patterns is often overlooked in these analyses [Citation5]. Studies have shown that media influence can significantly alter search behaviours and patterns [Citation14]. For evidence, studies of suicide have shown that televised depictions of suicide methods can lead to an increase in both suicides and suicide attempts by these methods [Citation15,Citation16]. Similarly, the development of media technology has not only broadened access to information about COVID-19 but has also shaped search behaviour. A recent study found an association between COVID-19 media coverage and polarisation of attitudes in the US [Citation17]. In addition, research suggests that social media, particularly Twitter, influences individuals’ attitudes and behaviour towards COVID-19 [Citation18]. As media coverage of COVID-19 increases, people may become more aware of the symptoms and more likely to seek information, driving up the volume of online searches [Citation19]. Therefore, it is vital to consider these factors, together with the impact of increased media coverage on symptom-related online searches, when analysing and using online search data in COVID-19 simulation models.
Furthermore, it is worth noting that a World Health Organisation (WHO) finding highlights the delayed onset of COVID-19 symptoms, with an incubation period of up to 14 days and subsequent infections affecting the following 14 days [Citation20]. In other words, the number of daily infections of each row in our dataset may influence the number of infection cases in the following 14 consecutive days. Additionally, there are 13 identified variants of SARS-CoV-2 in circulation, including notable strains such as Delta and Omicron, each with different rates of infection and transmissibility [Citation21,Citation22]. However, traditional machine learning and deep learning training methods respond to a randomly divided dataset to create a training and test set. The training set is used to build the model, while the test set is used to validate the model. Consequently, the traditional approach of randomly splitting datasets to train machine learning and deep learning-based simulation models does not provide reliable results in COVID-19 concurrent infection, as it is incapable of capturing the logical progression of the infection process. Recent studies have also confirmed the inadequacy of the traditional one-off training method for simulating COVID-19 infection cases [Citation23]. It is imperative to identify a new training method for the simulation model that reflects the infection and transmission patterns of COVID-19.
Therefore, this study has two main aims: (1) To introduce a novel machine learning-based approach to analyse the influence of comorbid conditions and media coverage on online symptom search data. Departing from conventional health-focused machine learning models that output a singular value denoting influence, our approach, leveraging quantile regression and Extreme Gradient Boosting (XG Boost), estimates a spectrum of influence. This provides a more dynamic and detailed perspective on the interplay between comorbid conditions, media impact and online searches for COVID-19 symptoms, ensuring a comprehensive analysis beyond simplistic numerical indicators. This approach allows the identification of search trends specifically attributable to individuals seeking information due to COVID-19 infection. (2) To develop a sophisticated recurrent neural network (RNN) model, enhanced with long short-term memory (LSTM) capabilities, using an innovative incremental batch learning training method. In this method, batches are created for each two-week period to accurately simulate the timeline of COVID-19 infection. This training approach will be designed to simulate COVID-19 infections in Australia and New Zealand, allowing continuous learning and adaptation to the varying infection rates and transmissibility characteristics of different COVID-19 variants. Meanwhile, the RNN-LSTM model will be enriched with the estimated range of influence of comorbid conditions and media coverage gathered from the first part of this study. This integration aims to create a more responsive and accurate model for simulating COVID-19 infection dynamics, capturing the nuanced effects of external factors on infection trends.
Overall, this approach will allow for more responsive and accurate modelling of pandemic progression in these regions. This paper also provides valuable insights into defining the COVID-19-specific component of the online symptom search and accurately estimating COVID-19 infection cases from online search data.
Materials and methods
Data sources
This study collected data from multiple sources for Australia and New Zealand over 30 weeks, spanning from August 1, 2022, to March 2, 2023. According to the information revealed by the United States Centres for Disease Control and Prevention (CDC) and the World Health Organisation, the symptoms caused by COVID-19 included fever, cough, sore throat, runny nose, headache, diarrhoea, etc [Citation24,Citation25]. Hence, this study extracts the search indexes for these symptoms from Google Trends [Citation26], which reflect the frequency of specific search terms relative to the total volume of searches on Google over a given period. In addition, the COVID-19 dataset on media coverage trends in Australia and New Zealand was obtained from MediaCloud [Citation27], an open-source media analysis platform that tracks, archives and makes searchable content from hundreds of newspapers and thousands of websites and blogs. In parallel, the COVID-19 case dataset is obtained from the John Hopkins University COVID-19 data repository [Citation28].
For the data pre-processing step, to process the online symptom search index, each symptom search index will be assigned a Symptom weight
with each symptom given an equal weight. Overall, the combined COVID-19 symptom search index
is defined as follows:
Impact of co-occurring illnesses with similar symptoms
To better estimate the online symptoms search led by COVID-19, the Google symptoms search index from the non-COVID period for Australia and New Zealand was first collected. Furthermore, the non-COVID period is from January 6, 2019 to October 10, 2019, based on the publicly available data.
The minimised impact of co-occurring illnesses on the symptoms search index is defined as follows:
Where the is a combined COVID-19 symptom search index from the data pre-processing step. The term
defined as the mean value of the symptoms search index during the non-COVID period, and the
is the daily COVID-19 symptom search.
Impact of media coverage
This study introduces a novel approach that goes beyond the scope of traditional health-focused machine learning models, which typically provide a single value indicating influence. Using a combination of quantile regression and Extreme Gradient Boosting (XG Boost), this approach uniquely estimates the upper and lower bounds of the impact of media reports on symptom search. This allows for a more dynamic and nuanced understanding of the factors driving online searches for COVID-19 symptoms, including the complex interplay between comorbid conditions and media coverage. This innovative approach provides a broader and more detailed perspective than traditional single-value analyses.
Assuming the daily COVID-19 symptom search, denoted consists of two components: the search behaviour of individuals who are infected with COVID-19 and seek advice via search engines (
), and the search behaviour of individuals who are influenced by media coverage (
). The weight of symptom searches influenced by media coverage (
) can be define as follows:
A quantile regression model was used to forecast the symptom search on day by using its current
and previous values
The loss function of the quantile regression-based model is defined as follows:
Where is a quantile level, and the range of
is from 0 to 1. The
is the predicted value of the symptom search index given prediction variables
and its previous values
The loss function facilitates the estimation of the residual error
at different quantile levels, using the symptom search index as a predictor variable. The
represents the unaccounted-for component in the symptom search, encompassing the impact of media reports and other unforeseen factors.
In this study, the quantile levels 10–90 and 25–75 are used to determine the maximum range of residual error and the quantile level 50 is used to determine the minimum range of residual error
The quantile level Q10 represents the estimated symptom search value below which 10% of the observations lie when the predictor variable, symptom search, is held constant. Similarly, the quantile level Q90 indicates the estimated symptom search value below which 90% of the observations lie when the predictor variable, symptom search, is held constant.
The equation provided serves as an example for quantile levels ranging from 10 to 90, specifically designed to define the maximum and minimum impact of media coverage.
Where represents the maximum unexplained component in symptom searches for quantile levels 10–90, determined by the difference between the residual errors at quantile level 90 (
) and those at quantile level 10
The
represents the minimum unexplained part, detemined by the residual errors at quantile level 50 (
).
Then, to estimate the effect of media coverage on symptom search, we used an Extreme Gradient Boosting (XG Boost) regressor model to estimate and
respectively, based on media coverage trends. The rationale for using the XG Boost regressor is similar to boosting logic in machine learning. Unlike the bagging approach, which trains models in parallel based on different datasets, the boosting model trains sequentially based on the residual error of the previously trained model. This approach is ideal for determining the impact of media coverage on the unexplained component in the quantile regression model. The XG boost model function is defined as:
Where is the initial prediction for dependent variables
or
at day
and
is the vector of weights assigned to each tree in the XG Boost model, with each weight indicating the strength of the corresponding tree’s prediction for a given observation.
is a hyperparameter that controls the strength of the regularisation term.
is the estimated maximum unexplained component using media coverage trends, while
is the estimated minimum unexplained component.
Lastly, the error ratio between the quantile regression model at the quantile 50 level and XG boost model was used to determine the range of the weight of symptom searches influenced by media coverage(
). The range of
is defined as follows:
As a result, the range of the symptom-seeking behaviour of infected individuals can be estimated and included as an additional attribute in the simulation model.
RNN-LSTM model for simulation for different quantiles
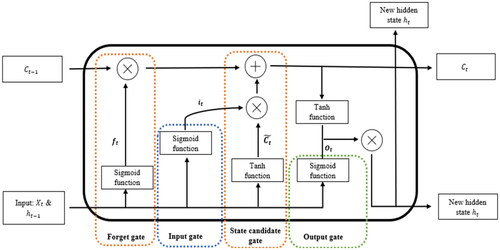
Moreover, the proposed method with Recurrent Neural Network (RNN)-Long Short-Term Memory (LSTM) embedding COVID-19 simulation model is established by using a novel incremental batch learning training method. In the incremental batch learning process, each batch is created for a 2-week period to simulate the COVID-19 infection period. During the training process, the first batch is used to train the model and initialise the model parameters. Then, the following batches are used for the incremental deep learning process to continuously update the model parameters to dynamically simulate COVID-19 infection cases. Overall, the proposed RNN-LSTM model provides a continuous learning capability driven by the training model, which allows the model to simulate the different infection rates and transmissibility of SARS-CoV-2 variants. and illustrate the LSTM architecture, where the input gate determines the proportion of new information to be added to the cell state, and the forget gate determines the proportion of information to be forgotten. The state candidate gate computes a new candidate cell state, and the output gate generates an output vector that weights the current cell state based on its relevance to the current output.
Table 1. Long Short-Term Memory Network (LSTM) formula.
To assess the performance of the proposed approach, it was compared with several machine learning methods using a traditional one-off training method based on the original symptom search index and the media coverage index. Overall, three performance metrics were used to evaluate the proposed model, including coefficient of determination (R2), root mean square error (RMSE) and mean absolute error (MAE).
Results
Minimised the impact of comorbid conditions with similar symptoms
presents the overall trend of symptom searches over a 30-week period in Australia and New Zealand, including both searches driven by co-occurring conditions with similar symptoms and those specifically attributed to COVID-19.
Figure 2. Minimised the impact of comorbid conditions with similar symptoms in (a) Australia and (b) New Zealand, respectively. The pink area represents the search range for comorbidities with similar symptoms. Specifically, the comorbid conditions with similar symptoms were estimated using data collected during the non-COVID-19 period, from 6th January 2019 to 10th October 2019. Meanwhile, the blue area in the graph represents the COVID-19 related search.
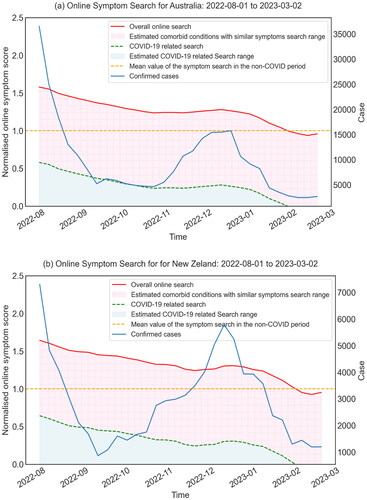
It was found that the estimated search score for COVID-19 related research in New Zealand increased slightly from 24.21% to 30.67% over the study period, while in Australia, it increased from 23.59% to 28.04%. These observations suggest a slightly higher propensity for COVID-19 related online information seeking among New Zealanders compared to their Australian counterparts. Notably, the increase in the search index is consistent with a marked increase in daily confirmed COVID-19 cases in Australia over the same period. A similar trend was also observed in New Zealand, where there was an increase in COVID-19 cases, particularly in November and December 2022.
Overall symptom search index remained higher than the mean value of the non-COVID period from August 2022 to February 2023, although it gradually decreased over the 30-week period in both countries. This change was mainly driven by the search index led by COVID-19.
Estimate the impact of media reports on the symptoms search
This section aims to evaluate the impact of media coverage and to determine the element of online symptom search using the proposed method by integrating both the media coverage index and the symptom search variables based on the 10–90 and 25–75 quantile scenarios.
As shown in , the pink shaded area represents the dynamic impact of media coverage on online symptom searches. This area is defined by the lower and upper bounds of media-influenced symptom search activity. This allows the examination of the variation of media influence during the study period. It was observed that the influence of media coverage was a dominant factor in the total online symptom searches. It also revealed a slightly greater impact of media coverage for individuals residing in New Zealand than Australia.
Figure 3. Estimated components of COVID-19 symptom search in Australia and New Zealand with (a) (c) the quantile 10–90 scenario, (b) (d) the quantile 25–75 scenario, respectively. The green area represents minimal media influence on symptom search, while the grey area represents minimal infected person-led symptom search. The pink area in the figure represents the uncertainty zone, bounded by the lower and upper bounds of media-influenced symptom searches, which visualises the dynamic impact of media coverage on online symptom searches and highlights the range of uncertainty in the analysis.
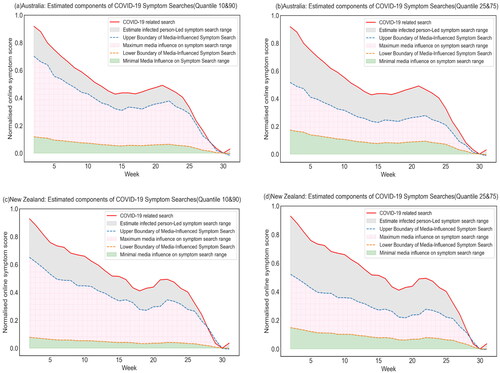
On average, the maximum effect of media coverage accounted for approximately 50.96% of online symptom searches in New Zealand (61.77% vs. 40.15%), while an average of 50.40% in Australia (63.43% vs. 37.37%). The study also showed that the impact of media coverage on people’s symptom-seeking behaviour increased slightly over the 30 weeks in both scenarios. The maximum impact of media influence on search, for the 10–90 quantile, the impact of media reports on people’s symptom-seeking behaviour peaks at around 0.10 in week 27 for Australia, while in New Zealand, it peaks at around 0.12 in week 30. A similar trend can be seen for the 25–75 quantile, with a peak of 0.04 at week 25 for Australia and a peak of 0.08 at week 30 for New Zealand.
For the estimated minimal effect of media influence, media coverage was found to have a stable and minimal effect on symptom searches for both countries. More specifically, the 50th percentile scenario is used to indicate the lowest possible impact of news reports on symptom searches for Australia and New Zealand. The margin of error for both countries was relatively constant, with only small fluctuations over the 30-week period. In Australia, the study found that the lower bound of media influence on symptom search accounted for approximately 12.94% of total symptom search in the 10–90 quantile scenario, whereas the lower bound of media influence on symptom search contributed approximately 18.90% in the 25–75 quantile scenario. Similar patterns were observed in New Zealand, where the lower bound of media-influenced symptom search contributed only 8.31% and 15.99%, respectively.
In addition, infected person-led symptom search accounted for approximately 23.63% of COVID-19 symptoms in Australia in the 10–90 quantile scenario, while it increased to approximately 43.73% of total symptom search in the 25–75 quantile scenario. Similar patterns were observed in New Zealand, where infected person-led symptom searches accounted for approximately 29.92% and 43.86% in the 10–90 and 25–75 quantile scenarios, respectively.
Proposed method with RNN-LSTM embedding model
Lastly, the estimated impact of comorbid conditions with similar symptoms and media reports were incorporated into the proposed method with the RNN-LSTM embedding model (quantile 10–90 & 25–75) for COVID-19 tracking analysis. We compared the proposed model (Quantile 10–90 & 25–75) with several simulation approaches, which were trained by using traditional methods in the online keyword search index domain. The results, presented in and , show a clear improvement in simulation performance using the proposed method with RNN-LSTM embedding (Quantile 10–90 & 25–75) for all three metrics. It is worth noting that the proposed method with RNN-LSTM embedding (quantile 25–75) achieves better results than RNN-LSTM embedding (quantile 10–90) for both Australia and New Zealand.
Figure 4. Estimation of COVID-19 cases using the proposed method with the RNN-LSTM embedding model for (a) Australia and (b) New Zealand for the 10–90 and 25–75 quantile scenarios.
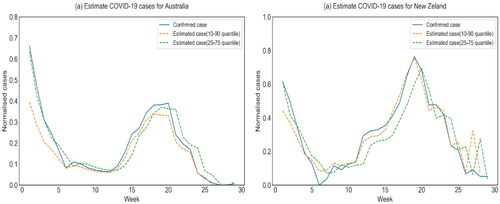
Table 2. Results of model evaluation.
Discussion
This study highlights the significant influence of media coverage and comorbid conditions with similar symptoms on online symptom searches. The proposed method, which integrates an RNN-LSTM embedding model, showed improvements over other simulation techniques, particularly after accounting for the impact of media coverage and the presence of comorbidities.
Regarding the online search data, the approach used in this study has successfully measured the magnitude of the impact of media coverage on symptom search using a machine learning-based component analysis model. It has been identified that most symptom search volume in Australia and New Zealand is driven by media reports and co-occurring illnesses, overshadowing the volume of symptom searches conducted by those infected with COVID-19. Previous studies have indeed found a statistically significant correlation between online interest and COVID-19 case numbers in various regions, including Europe [Citation29] (specifically Italy, Spain, France, the UK and Germany), the USA and China [Citation3,Citation30]. However, these studies do not adequately take into account the impact of co-occurring conditions with similar symptoms, a factor that our research has significantly identified. This study has shown that symptom searches for co-occurring conditions account for a larger proportion of online symptom searches than those specifically related to COVID-19. This suggests that using online symptom search data directly in COVID-19 infection simulations may lead to inaccurate or unreliable results, given the significant inclusion of symptom searches related to other diseases.
In addition to the impact of co-occurring illnesses with similar symptoms, the role of media coverage in online COVID-19 symptom search has been previously primarily ignored in simulation models [Citation4,Citation30]. However, a recent study has attempted to use an auto-regression model to estimate the influence of media reports on COVID-19 symptom search [Citation5]. However, the constant weight given to the estimation of symptom searches influenced by news reports in the auto-regression model contradicts our finding, as shown in the component analysis of this study, that the influence of media coverage on online symptom searches is dynamic and fluctuates over time. Therefore, the auto-regression model does not accurately capture the fluctuating impact of media coverage on individuals’ search behaviour. Meanwhile, the proposed simulation model outperformed their ElasticNet model, which was previously used for COVID-19 simulation modelling.
In the context of simulation modelling, it is crucial to thoroughly analyse and understand the dynamics of virus transmission and infection patterns. By monitoring changes in transmission and infection estimates over time, valuable insights into epidemiological situations can be gained, as well as an assessment of the effectiveness of implemented outbreak control measures [Citation31,Citation32]. Researchers have identified an asymptomatic period of between 7 and 14 days for each infected individual [Citation20,Citation33]. This phenomenon poses a significant challenge to pandemic control, as these individuals are unknowingly contributing to the transmission of the virus. In light of these factors, the traditional method of randomly splitting datasets, which is commonly used for simulation purposes, proves ill-suited to accurately represent the complex dynamics of COVID-19 transmission. An alternative approach, incremental batch learning, emerges as a more viable option. This method allows for continuous learning and adaptation of the simulation model to capture the evolving infection and transmission patterns better, providing greater accuracy and reliability in simulating COVID-19 scenarios.
Moreover, the Recurrent Neural Network (RNN)-Long Short-Term Memory (LSTM) has been identified as a reliable approach for handling long sequential time series data, as supported by previous research [Citation23]. The Long Short-Term Memory Network (LSTM), which combines the RNN with effective solutions for short-term memory limitations and gradient problems, is a promising technique [Citation34]. Notably, both proposed methods using the RNN-LSTM embedding model (quantile 25–75 and quantile 10–90) successfully simulate the spread of COVID-19 in Australia and New Zealand, as shown in this study. Interestingly, the quantile 25–75 model outperforms the quantile 10–90 model. This study suggests that the differences in prediction accuracy are primarily influenced by the maximum impact of media influence on these searches. Specifically, the estimated maximum impact of media influence on searches decreases from 73.23% in the 10–90 quantile range to 56.21% in the 25–75 quantile range.
Overall, the proposed method provides valuable insights into the factors contributing to online symptom search and highlights the significant impact of media coverage in both the 10–90 and 25–75 quantile scenarios. Furthermore, this approach can inform the response to COVID-19 by enabling governments to estimate the approximate number of infected cases through online symptom searches.
This study is not without some limitations. Firstly, our study was limited to Australia and New Zealand, a geographical limitation that may limit the generalisability of our results to other regions. This focus raises concerns about the external validity of our findings, as infection cases and trends outside these countries were not considered. Secondly, our analysis relied primarily on the Google online symptom search dataset, potentially overlooking the diversity of search behaviour across different search engines. The reliance on a single search engine may not capture the full range of online behaviour related to COVID-19 symptoms, thereby affecting the representativeness of our dataset and potentially introducing bias. Thirdly, the digital divide and different levels of internet access in diverse populations pose a further challenge. Relying on online search data risks excluding groups with limited internet access or who are less likely to use online search engines, particularly in regions with lower digital connectivity. These limitations highlight the need for a more comprehensive approach in future research to ensure wider validity and inclusivity.
Conclusion
In summary, this study highlights that in Australia and New Zealand, symptom searches influenced by media reports and co-morbidities significantly outweigh the search behaviour of COVID-19 infected individuals seeking advice via search engines. This suggests that direct use of online symptom search data in COVID-19 infection models could lead to inflated infection estimates. This study shows that media-influenced searches are slightly more common in New Zealand than Australia.
Furthermore, an RNN-LSTM model with innovative incremental batch learning, was used to dynamically simulate the varying infection rates and transmissibility of SARS-CoV-2 variants. Enhanced with estimated ranges of influence from comorbid conditions and media coverage, this model provides a more adaptive and accurate tool for simulating COVID-19 infection dynamics, effectively capturing the complex effects of external factors on infection trends. The model demonstrates a clear improvement in three key metrics(R2, RMSE and MAE) and effectively manages the impact of comorbid conditions on symptom search using historical data from the non-COVID-19 period.
Overall, the approach used in this study provides novel insights into the estimation of COVID-19 infections through online symptom searches. This approach could assist governments in accurately estimating COVID-19 cases, thereby improving public health surveillance and response strategies.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- World Health Organization. n.d. Weekly epidemiological update on COVID-19 - 1st March 2023. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19–-1-march-2023.
- Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin Immunol. 2020;215:108427. doi:10.1016/j.clim.2020.108427.
- Mavragani A, Gkillas K. COVID-19 predictability in the United States using google trends time series. Sci Rep. 2020;10(1):20693. doi:10.1038/s41598-020-77275-9.
- Higgins TS, Wu AW, Sharma D, et al. Correlations of online search engine trends with coronavirus disease (COVID-19) incidence: infodemiology study. JMIR Public Health Surveill. 2020;6(2):e19702. doi:10.2196/19702.
- Lampos V, Majumder MS, Yom-Tov E, et al. Tracking COVID-19 using online search. NPJ Digit Med. 2021;4(1):17. doi:10.1038/s41746-021-00384-w.
- Strzelecki A. The second worldwide wave of interest in coronavirus since the COVID-19 outbreaks in South Korea, Italy and Iran: a google trends study. Brain Behav Immun. 2020;88:950–951. doi:10.1016/j.bbi.2020.04.042.
- Walker A, Hopkins C, Surda P. Use of google trends to investigate loss‐of‐smell–related searches during the COVID‐19 outbreak. Int Forum Allergy Rhinol. 2020;10(7):839–847. doi:10.1002/alr.22580.
- Rabiolo A, Alladio E, Morales E, et al. Forecasting the COVID-19 epidemic by integrating symptom search behavior into predictive models: infoveillance study. J Med Internet Res. 2021;23(8):e28876. doi:10.2196/28876.
- Cai O, Sousa-Pinto B. United States influenza search patterns since the emergence of COVID-19: infodemiology study. JMIR Public Health Surveill. 2022;8(3):e32364. doi:10.2196/32364.
- Wojcik S, Bijral AS, Johnston R, et al. Survey data and human computation for improved flu tracking. Nat Commun. 2021;12(1):194. doi:10.1038/s41467-020-20206-z.
- Huang DC, Wang JF, Huang JX, et al. Towards identifying and reducing the bias of disease information extracted from search engine data. PLoS Comput Biol. 2016;12(6):e1004876. doi:10.1371/journal.pcbi.1004876.
- Flu (influenza). Healthdirect; 2023 Apr 20. https://www.healthdirect.gov.au/flu.
- Influenza. Ministry of Health NZ. n.d. https://www.health.govt.nz/your-health/conditions-and-treatments/diseases-and-illnesses/influenza.
- Marlow CA. The structural determinants of media contagion [doctoral dissertation]. Cambridge: Massachusetts Institute of Technology; 2005.
- Gould M, Jamieson P, Romer D. Media contagion and suicide among the young. American Behav Sci. 2003;46(9):1269–1284. doi:10.1177/0002764202250670.
- Pescara-Kovach L, Raleigh MJ. The contagion effect as it relates to public mass shootings and suicides. J Campus Behav Intervention. 2017;5:35–45.
- Hart PS, Chinn S, Soroka S. Politicization and polarization in COVID-19 news coverage. Sci Commun. 2020;42(5):679–697. doi:10.1177/1075547020950735.
- Bridgman A, Merkley E, Loewen PJ, et al. The causes and consequences of COVID-19 misperceptions: understanding the role of news and social media. HKS Misinfo Rev. 2020;1(3). doi:10.37016/mr-2020-028.
- Krawczyk K, Chelkowski T, Laydon DJ, et al. Quantifying online news media coverage of the COVID-19 pandemic: text mining study and resource. J Med Internet Res. 2021;23(6):e28253. doi:10.2196/28253.
- World Health Organization. Health topics. Available from: https://www.who.int/health-topics/
- Australian Government Department of Health and Aged Care. COVID-19 disease and symptoms. Available from: https://www.health.gov.au/health-alerts/covid-19/symptoms
- Yan L, Talic S, Wild H, et al. Transmission of SARS-CoV-2 in a primary school setting with and without public health measures using real-world contact data: a modelling study. J Global Health. 2022;12:05034. doi:10.7189/jogh.12.05034.
- Lyu S, Adegboye O, Adhinugraha K, et al. COVID-19 prevention strategies for Victoria students within educational facilities: an AI-based modelling study. Healthcare. 2023;11(6):860. doi:10.3390/healthcare11060860.
- World Health Organization. n.d. Coronavirus. https://www.who.int/health-topics/coronavirus#tab=tab_3.
- Centers for Disease Control and Prevention. n.d. Symptoms of COVID-19. https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html.
- Google. n.d. Google trends. https://trends.google.com/home.
- Media cloud. Media Cloud. n.d. https://search.mediacloud.org/.
- CSSEGISandData. n.d. CSSEGISANDDATA/covid-19: novel coronavirus (COVID-19) cases, provided by JHU CSSE. GitHub. https://github.com/CSSEGISandData/COVID-19.
- Mavragani A. Tracking COVID-19 in Europe: infodemiology approach. JMIR Public Health Surveill. 2020;6(2):e18941. doi:10.2196/18941.
- Saegner T, Austys D. Forecasting and surveillance of COVID-19 spread using google trends: literature review. Int J Environ Res Public Health. 2022;19(19):12394. doi:10.3390/ijerph191912394.
- Wu JT, Leung K, Bushman M, et al. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med. 2020;26(4):506–510. doi:10.1038/s41591-020-0822-7.
- Adly AS, Adly AS, Adly MS. Approaches based on artificial intelligence and the internet of intelligent things to prevent the spread of COVID-19: scoping review. J Med Internet Res. 2020;22(8):e19104. doi:10.2196/19104.
- Liu Z, Magal P, Seydi O, et al. Predicting the cumulative number of cases for the COVID-19 epidemic in China from early data. arXiv preprint arXiv:2002.12298. 2020.
- Zarzycki K, Ławryńczuk M. LSTM and GRU neural networks as models of dynamical processes used in predictive control: a comparison of models developed for two chemical reactors. Sensors. 2021;21(16):5625. doi:10.3390/s21165625.