Abstract
The complete mitochondrial genome, containing 17,526 bp, was determined from the pitviper Bothrops jararaca. It is the first mitogenome for the most medically important genus of snake in Latin America. This mitogenome has common snake mitochondrial features such as a duplicated control region that has nearly identical sequences at two different locations of the mitogenome and a translocation of tRNA-Leu (UUR). Besides, we found a translocation of the tRNA-Pro compared to Colubridae snakes. Finally, an unusual possible duplication containing a tRNA-Phe was observed for the first time and may represent a marker of the genus.
Bothrops jararaca belongs to Viperidae family and is distributed mainly in the Southeast and South Regions of Brazil. It is the most medically important snake in South America, and its venom is one of the most studied among all snakes (Cardoso Citation1990, Goncalves-Machado et al. Citation2015). Distinct populations are characterized by a high genetic diversity, whereas sister species such as B. insularis and B. alcatraz have some genetic proximity to certain populations (Grazziotin et al. Citation2006). However, no complete mitochondrial genome is available for the genus, which encompasses up to 44 species, most of them of high medical interest.
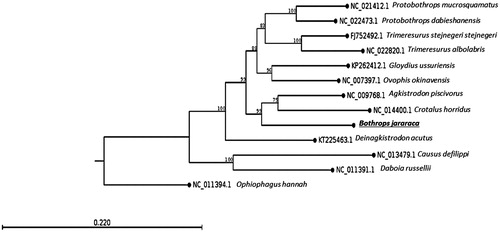
Here, we determined the complete mitogenome of B. jararaca (GenBank accession number KU194299) by whole genome shotgun using Illumina paired-end sequencing (Illumina HiSeq1500). The specimen was collected at Embu das Artes, São Paulo State, Brazil – Geospatial coordinates: −23.654919, −46.864827 – and is disposed at the Herpetological Collection of the Special Laboratory of Zoological Collections from Butantan Institute (accession number: IBSP 84406). The genes were predicted by two independent softwares (MITOS and DOGMA) and were manually curated. A phylogenetic tree with the closest species available is shown in . The gene order and structure is similar to that found in the family (Dong & Kumazawa Citation2005; Huang et al. Citation2014). The total length of this mitogenome is 17,526 bp and it contains 13 protein-coding genes (PCGs), 2 rRNA genes, 23 tRNA genes (including a possible new tRNA which has 88% of sequence similarity to tRNA-Phe), and 2 control regions. Most genes are coded on the heavy strand (H-strand) except for the ND6 and 8 tRNAs. The base composition of the light strand is 31.9% A, 24.5% T, 13.3% G, and 30.3% C. The high AT content agrees with that of other snakes (Jang & Hwang Citation2011; Li et al. Citation2014).
Figure 1. The maximum-likelihood phylogenetic tree of B. jararaca mitocondrial DNA and 11 closely related species (Viperidae) whose mitogenome sequences have been reported, indicates the placement of B. jararaca genome in the Crotalinae subfamily, closely to the sister taxa (Crotalus and Agkistrodon) of genus Bothrops. The tree was generated using 1000 replicates and the boostrap values are shown on each node. The mitogenome of Ophiophagus hannah (Elapidae) was used as an outgroup.
The mitogenome of B. jararaca contains almost identical duplicate regions corresponding to the Control Regions (CRs). CRI is located between tRNA-Thr and tRNA-Phe genes and CRII is located between the tRNA-Pro and tRNA-Leu genes. As in other snakes, the tRNA-Leu (UUR) gene, which occurs between the 16S rRNA and ND1 genes in other vertebrates, is located downstream of the CRII. As in other Viperidae snakes (Jiang et al. Citation2007; Hall et al. Citation2013; Huang et al. Citation2014), the tRNA-Pro gene is located between CRII and tRNA-Ile gene instead of the CRI 5′ vicinity (a pseudo tRNA-Pro gene can be found between CRII and tRNA-Ile gene in Colubridae) (Jang & Hwang Citation2011). The predicted origin of L-strand replication (OL) is 35 bp in length and is located between the tRNA-Asn and tRNA-Cys.
A distinctive feature in the B. jararaca mitogenome is the presence of a possible tRNA-Phe duplication, with 88% identity. The tRNA-Phe-like gene is found downstream of the CRI region. We consider the tRNA-Phe coded in the position 1 to 65 as the functional one; the role of the other one needs to be determined. It will remain to be investigated if this distinctive feature is conserved among other related species, but if it is, the synapomorphy may be a useful character to help in elucidating the controversial phylogeny of the group.
This work presents the first mitogenome of a Bothrops species, showing that it has both similar and distinctive features when compared to those from other genera, providing additional molecular data for phylogenetic studies of snakes.
Disclosure statement
The authors have declared that no conflict of interest exists.
Additional information
Funding
References
- Cardoso J. 1990. Bothropic accidents. Mem Inst Butantan. 52:43–44.
- Dong S, Kumazawa Y. 2005. Complete mitochondrial DNA sequences of six snakes: phylogenetic relationships and molecular evolution of genomic features. J Mol Evol. 61:12–22.
- Goncalves-Machado L, Pla D, Sanz L, Jorge RJ, Leitao-De-Araujo M, Alves ML, Alvares DJ, De Miranda J, Nowatzki J, De Morais-Zani K, et al. 2015. Combined venomics, venom gland transcriptomics, bioactivities, and antivenomics of two Bothrops jararaca populations from geographic isolated regions within the Brazilian Atlantic rainforest. J Proteomics. 135:73–89.
- Grazziotin FG, Monzel M, Echeverrigaray S, Bonatto SL. 2006. Phylogeography of the Bothrops jararaca complex (Serpentes: Viperidae): past fragmentation and island colonization in the Brazilian Atlantic Forest. Mol Ecol. 15:3969–3982.
- Hall JB, Cobb VA, Cahoon AB. 2013. The complete mitochondrial DNA sequence of Crotalus horridus (timber rattlesnake). Mitochondrial DNA. 24:94–96.
- Huang X, Zhang L, Pan T, Zhang B. 2014. Mitochondrial genome of Protobothrops dabieshanensis (Squamata: Viperidae: Crotalinae). Mitochondrial DNA. 25:337–338.
- Jang KH, Hwang UW. 2011. Complete mitochondrial genome of the black-headed snake Sibynophis collaris (Squamata, Serpentes, Colubridae). Mitochondrial DNA. 22:77–79.
- Jiang ZJ, Castoe TA, Austin CC, Burbrink FT, Herron MD, Mcguire JA, Parkinson CL, Pollock DD. 2007. Comparative mitochondrial genomics of snakes: extraordinary substitution rate dynamics and functionality of the duplicate control region. BMC Evol Biol. 7:123.
- Li E, Feng D, Yan P, Xue H, Chen J, Wu XB. 2014. The complete mitochondrial genome of Oocatochus rufodorsatus (Reptilia, Serpentes, Colubridae). Mitochondrial DNA. 25:449–450.

