Abstract
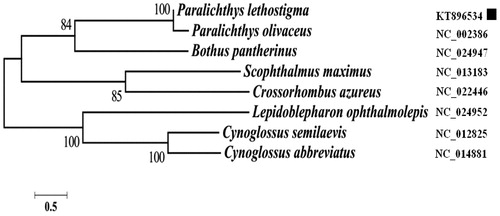
In this study, the complete mitochondrial genomic sequence of southern flounder Paralichthys lethostigma was determined. The circular mitogenome is 16 843 bp in length and encodes 37 typical animal mitochondrial genes and contains a putative D-loop region. Phylogenetic analysis based on the mitochondrial genomes of southern flounder indicated that P. lethostigma and P. olivaceus are the most closely related species, which strongly supports their close phylogenetic affinity.
The southern flounder, Paralichthys lethostigma, is a large, ecologically and commercially important benthic flatfish that inhabits the coastal waters from North Carolina, USA to northern Florida, and from Tampa Bay, Florida along the Gulf coast into southern Texas (Froeschke et al. Citation2011). Since the southern flounder was introduced into China from North Carolina in 2002, it has shown great promise as an aquaculture species (Xu et al. Citation2013). The successful management of the southern flounder fishery depends on a comprehensive understanding of its biology, including its population genetic structure. However, it has received little attention in genomic or population structure studies. Molecular markers, including those in the mitochondrial genome, can be used to provide insight into the population diversity and evolution of species (Schuster Citation2008).
Ten individuals of P. lethostigma were collected from the Center for Marine Science, University of North Carolina Wilmington, NC, USA (34°08′N, 77°52′E) and stored in Yellow Sea Fisheries Research Institute Fish reproduction center, Qingdao, China (number as YSFRI-Paralichthys-lethostigma-shi-SF1). The whole genomic DNA was extracted from muscle tissues and the mitogenome was amplified with universal metazoan primer pairs and completely sequenced with a 3730 × l DNA Analyzer (Fan et al. Citation2012). The complete mitochondrial genome of P. lethostigma has a length of 16 843 bp (GenBank accession no. KT896534). It encodes 13 protein-coding genes, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and one putative control region (D-loop), as in other vertebrates (Miya et al. Citation2001). Two kinds of start codons were identified in the 13 protein-coding genes, the typical ATG codon in most genes and GTG in the COI gene. Six protein-coding genes (ND1, COI, ATPase 8, ND4L, ND5 and ND6) end with complete termination codons (TAA or TAG), whereas seven terminate in an incomplete stop codon TA (ND2, ATPase 6 and COIII) or T (COII, ND3, ND4 and CytB), which is completed (to TAA) by posttranscriptional polyadenylation (Ojala et al. Citation1981). Three overlapping reading frames are present, ATP8–ATP6, ND4L–ND4 and ND5–ND6, which overlap by 8, 5 and 2 nucleotides, respectively. The 12S and 16S rRNA genes are 949 and 1731 bp long, respectively, and are located between the tRNAPhe and tRNALeu(UAA) genes and are separated by tRNAVal. The 22 tRNA genes are interspersed throughout the genome, ranging in size from 68 to 73 bp, and the gene products fold into cloverleaf secondary structures. The putative control region is located between tRNAPhe and tRNAPro, with a length of 1120 bp. Phylogenetic analysis based on the eight complete mitochondrial genome data of pleuronectiformes show that P. lethostigma belongs to a paralichthyidae clade and is closely related to P. olivaceus ().
Disclosure statement
This study was supported by the National High Technology Research and Development Program of China (2012AA10A413), the National Natural Science Foundation of China (31201982), the Shandong Youth Scientist Awards Foundation (BS2013SW042) and the China Agriculture Research System (CARS-50). All the authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Fan X, Qiao HJ, Xu D, Cao SN, Zhang XW, Wang YT, Mou SL, Ye NH. 2012. Complete mitochondrial genome of the sea slug, Placida sp., contains unusual noncoding regions Mitochondr DNA. 24:219–221.
- Froeschke BF, Sterba-Boatwtight B, Stunz GW. 2011. Assessing southern flounder (Paralichthys lethostigma) long-term population trends in the northern Gulf of Mexico using time series analyses. Fish Res. 108:291–298.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18:1993–2009.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Schuster SC. 2008. Next-generation sequencing transforms today's biology. Nat Methods. 5:16–18.
- Xu YJ, Liao MJ, Wang YG, Liu ZC, Qin P, Zhang Z, Rong XJ, Liu XZ. 2013. Isolation of novel microsatellite markers from Paralichthys lethostigma (Paralichthyidae) and their cross-species application in Pleuronectiformes. Genet Mol Res. 12:6767–6772.