Abstract
The complete mitochondrial DNA of biraphid benthic diatom, Navicula ramosissima TA439 was sequenced and characterized. The circular mitogenome contains 67 genes in 48,652 bp (31.1% GC), including 41 protein-coding, 24 transfer RNA (tRNA) and 2 rRNA genes. Twenty-four protein-coding sequences (CDS, 59%) have start with ATG codon and 17 CDS start with alternatives such as ATA (5), ATT (6), TTA (5) and TTG (1). The GC content of tRNA genes (42.1%) is relatively higher than those of the rRNA (35.2%) and CDS (30.5%). Three genes are consisted of multiple exons and introns, i.e. cox1 (three exons, two introns), rps11 (two exons, one intron), rrl (four exons, three introns). Phylogeny of diatoms based on mitogenome data (34 CDS, 8530 amino acids combined) supports the monophyly of Naviculales, including N. ramosissima (Naviculaceae), Berkeleya fennica (Berkeleyaceae), Fistulifera solaris (Stauroneidaceae) and Phaeodactylum tricornutum (Phaeodactylaceae). Mitogenome data may be useful for phylogenetic study of the diatoms and stramenopiles.
Navicula ramosissima (C.Agardh) Cleve is ubiquitous species in brackish and marine water, and is dominant during the winter months (Werner Citation1977; Snoeijs & Kautsky Citation1989). The species is included in biraphid motile diatom because individual cell moves and grows colonially in branched mucilaginous tube (Lobban & Navarro 2013). The species is one of the most numerous diatoms in tidal mudflat of the west coast of Korea, thus subject to primary production monitoring researches. However, no mitochondrial data available yet, only nuclear rRNAs (18S and 28S) and plastid genes (rbcL and 16S rRNA) were reported to public. In the present study, we determined novel mitogenome of N. ramosissima TA439 isolated from the Geunsoman, Taean (36°44′12.06″ N 126°10′47.52″ E), Korea (strain available upon request). We followed next-generation sequencing strategy as described in An et al. (Citation2016) and Yang et al. (Citation2015).
The complete mitogenome (mtDNA) of N. ramosissima TA439 (GenBank accession no. KX343079) indicates a 48,652 bp of circular genome with 31.1% GC content. Total 67 genes are encoded in 40,335 bp (83% of total size) exclude introns, including of 41 protein-coding sequences (CDS; i.e. 18 respiratory, 16 ribosomal and 7 hypothetical protein-coding genes; total 34,236 bp with 31.5% GC), 2 ribosomal RNA (rrl and rrs; total 4286 bp exclude introns with 35.2% GC) genes for large and small subunits, and 24 transfer RNA (tRNA; total 1813 bp with 42.1% GC) genes. The ATG is the most common start codon used in 24 protein-coding sequences (CDS; 59% of total CDS) and followed by ATT (six CDS), ATA (five CDS), TTA (five CDS), and TTG (one CDS) codons. All CDS use the stop codon either TAA (31 CDS; 76%) or TAG (10 CDS). Three genes include multiple exons and introns, i.e. cox1 (three exons and two introns), rps11 (two exons, one intron), rrl (four exons, three introns). Two cox1 introns (2439 bp, 2325 bp) harbour intronic ORF such as ORF717 and ORF750. The intron-2 (2422 bp) and intron-3 (2355 bp) of rrl introns harbour ORF558 and ORF218, respectively. No intronic ORF encoded in rrl intron-1 (834 bp) and rps11 intron (37 bp). All intronic ORF matched with diatoms’ intronic ORF which encoded the Group II intron-specific reverse transcriptase/muturase, e.g. orf79 of Pseudo-nitzschia multiseries (GI: 836614283). The 24 tRNA genes, ranging from 69 to 89 bp in length, show typical cloverleaf secondary structures.
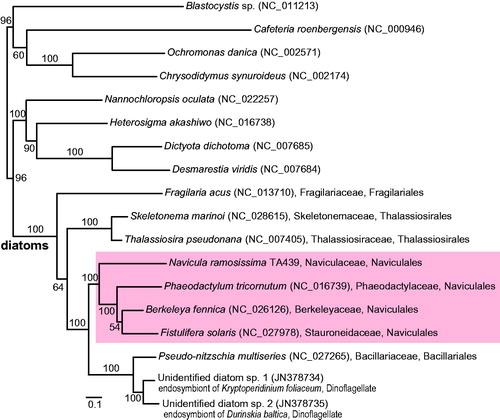
Translated amino acid sequences of N. ramosissima TA439 mitogenome used for phylogenetic analyses with those of all available diatoms, i.e. the best tree construction and bootstrap analysis under the maximum-likelihood (ML) criterion. The best phylogeny of 34 CDS concatenated data (8530 amino acids) supports the monophyly of Bacillariophyta (100% ML bootstrap support, MLB; ) and congruent with previous study (An et al. Citation2015). Naviculales is grouped with Bacillariales within Bacillariophycidae (100% MLB). Monophyletic Naviculales (100% MLB) included N. ramosissima (Naviculaceae), B. fennica (Berkeleyaceae), F. solaris (Stauroneidaceae), and P. tricornutum (Phaeodactylaceae). New phylogeny suggested that additional mitogenome data from representative lineages may be useful for the systematic study of diatoms.
Figure 1. ML tree of the diatoms based on the mitochondrial genome data, 34 CDS concatenated data (8530 amino acids). The support value of each node calculated from ML bootstrap analysis, 100 non-parametric bootstrapping. Diatom species name were followed by the GenBank accession number in parenthesis and family and order names.
Acknowledgements
We thank Mr. Dong Min Park for the assistance of sample collection.
Disclosure statement
The authors are responsible for the content and writing of the paper. The authors report no conflicts of interest.
Funding information
This work was supported by grants from the Korea Institute of Ocean Science & Technology (PE99412) to JHN and Marine Biotechnology Program (PJT200620) funded by the Ministry of Oceans and Fisheries, Korea to ECY.
References
- An SM, Kim SY, Noh JH, Yang EC. 2015. Complete mitochondrial genome of Skeletonema marinoi (Mediophyceae, Bacillariophyta), a clonal chain forming diatom in the west coast of Korea. Mitochondrial DNA. DOI: 10.3109/19401736.2015.1106523. [Epub ahead of print].
- An SM, Noh JH, Choi DH, Lee JH, Yang EC. 2016. Repeat region absent in mitochondrial genome of tube-dwelling diatom Berkeleya fennica (Naviculales, Bacillariophyceae). Mitochondrial DNA Part A. 27:2137–2138.
- Lobban CS, Navarro JN. 2013. Gato hyalinus gen. et sp. nov., an unusual araphid tube-dwelling diatom from Western Pacific and Caribbean islands. Phytotaxa. 127:22–31.
- Snoeijs PJM, Kautsky U. 1989. Effects of ice-break on the structure and dynamics of a benthic diatom community in the northern Baltic Sea. Bot Mar. 32:547–562.
- Yang EC, Kim KM, Kim SY, Lee J, Boo GH, Lee J-H, Nelson WA, Yi G, Schmidt WE, Fredericq S, et al. 2015. Highly conserved mitochondrial genomes among multicellular red algae of the Florideophyceae. Genome Biol Evol. 7:2394–2406.
- Werner D. 1977. The biology of diatoms. vol. 13. Berkeley and New York: University of California Press. pp. 498.

