Abstract
The complete mitochondrial genome of the Wuchuan Odorous Frog was 18,256 bp in length including 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and a control region and was similar to that of typical vertebrates. The base composition was 27.89% A, 29.00% C, 15.34% G, and 27.78% T. All genes were encoded on the H-strand except ND6 and eight tRNA genes (tRNAPro, tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, and tRNAGlu), which were encoded on the L-strand. The phylogenetic relationship of Anura based on complete mitochondrial genomes showed that O. wuchuanesis is closest to O. margaretae with strong support and the genetic distance between Ranidae, Dicroglossidae, and Rhacophoridae was closer than others.
The wuchuan odorous frog (Odorrana wuchuanensis) belongs to the order Anura, the Ranidae and is an endemic species to China with a narrow distribution in karst area in southwestern China. The number of O. wuchuanensis is decreasing due to the increasing attenuation of habitat. In this study, we determined the complete mitochondrial DNA (mtDNA) sequence of O. wuchuanensis (Genbank accession: KU680791) for further evolutionary researches as well as for the protectionof this species.
The toe-clip tissue of O. wuchuanensis were collected from a specimen (Sample No. LBML 5230) stored in MAO LAN national nature reserve (N25°09′∼25°20′, E107°52′∼108°45′) in Libo county, Guizhou province, China. Total genomic DNA was extracted with the TIANamp Genomic DNA Kit (Tiangen, Beijing, China) following the kit’s instructions. The complete mitochondrial genome was amplified by polymerase chain reaction (PCR) with eight primers, of which primer L1 and Cytb is available from references (Rassmann Citation1997; Zhang et al. Citation2013) while primer ND2 and ND4 was designed based on the complete mtDNA sequence of O. margaretae (NC_024603) and the others were designed specifically for the rest long fragments of mtDNA amplified in long and accurate PCR (LA-PCR). All the PCR fragments were directly sequenced.
The complete sequence of the mitogenome of O. wuchuanensis is 18,256 bp in length with 55.66% A + T content. The overall nucleotide composition of this genome was 27.89% A, 29.00% C, 15.34% G, and 27.78% T, similar to typical vertebrate mtDNA. The genome contained 13 protein-coding genes, 2 ribosomal RNA genes, 22 tRNA genes, and 1 control region. The total length of 22 tRNA genes was 1527 bp, ranging from 64 bp (tRNACys) to 74 bp (tRNALeu(UUR)). The total length of 13 protein-coding genes was 11,301bp, of which all genes were encoded on the H-strand except ND6, as well as eight tRNA genes (tRNAPro, tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu), which were encoded on the L-strand. The start codons were ATG for ND1, COII, ATP8, COIII, ND3, ND4L, ND4, ND5, ND6, and CYTB while ATT for ND2, GTG for COI and ATA for ATP6. COI and ND6 terminated with AGG as stop codon; ATP8, ND4L, ND4 with TAA; ND5, CYTB with TAG, and other six protein-coding genes end with an incomplete stop codon (a single stop nucleotide T). The two ribosomal RNA genes including 12SrRNA (1219 bp in length) and 16SrRNA (2874 bp in length) were commonly located between tRNAPhe and tRNALeu(UUR). The control region was 2857 bp in length with 61.95% A + T content.
Within the complete mitochondrial genome of O. wuchuanensis, there were five reading frame overlaps (The tRNAGln and tRNAMet share two nucleotides; OL and tRNACys share three nucleotides; COI and tRNASer share nine nucleotides; ATP8 and ATP6 share four nucleotides; ND4L and ND4 share seven nucleotides). Compared with the Green Odorous Frog (O. margaretae) (Chen et al. Citation2015) and Concave-eared Torrent Frog (Amolops tormotus) (Su et al. Citation2007), all of the start codons were similar except ND1, ND2, COI, and ND3. The stop codon was GTG for ND1 in O. margaretae, while ATG in O. wuchuanensis and A. tormotus. The stop codon was ATG for ND2 in A. tormotus while ATT in O. wuchuanensis and O. margaretae. The stop codon was ATG for COI in A. tormotus while GTG in O. wuchuanensis and O. margaretae. The stop codon was GTG for ND3 in O. margaretae while ATG in O. wuchuanensis and A. tormotus.
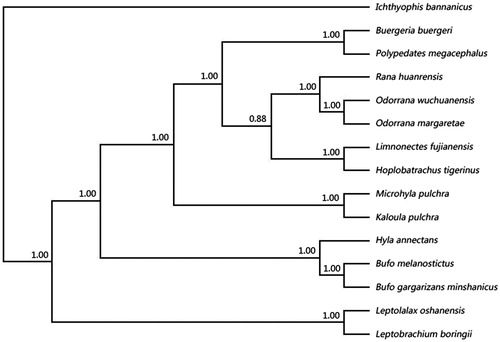
The phylogenetic analysis of the Anura based on complete mitochondrial genomes was conducted with Bayesian inference (BI). The best-fitted substitution model was determined in MODELTEST v.3.7 (Posada & Crandall Citation1998) by Akaike Information Criterion (AIC). The BI was conducted in MrBayes 3.2 (Ronquist et al. Citation2012) with Markov Chain Monte Carlo (MCMC) running for 50,00,000 generations, and sampling frequency for 100. A consensus tree was calculated after omitting the first 25% trees as burn-in. The 50% majority rule consensus tree inferred from Bayesian analysis with high clade credibility values shows that the Leptolalax oshanensis and Leptobrachium boringii are grouped as the sister taxon to a clade comprising the others of Anura (). Specially, O. wuchuanensis and O. margaretae are closest and they are grouped as the sister taxon to a clade comprising Rana huanrensis. Besides, the genetic distance between Ranidae, Dicroglossidae, and Rhacophoridae was closer than others in phylogenetic relationship as shown in .
Figure 1. It is the 50% majority rule consensus tree from the Bayesian inference (BI) of complete mtDNAs. The values above branches represent Bayesian clade credibility values. The phylogenetic tree contains 14 species of Anura and Ichthyophis bannanicus (NC_006404) used as outgroup. Sequence data used in this study were the following: Hoplobatrachus tigerinus (AP011543), Limnonectes fujianensis (AY974191), Bufo gargarizans minshanicus (KM587710), Bufo melanostictus (AY458592), Kaloula pulchra (AY458595), Microhyla pulchra (KF798195), L. boringii (KJ630505), L. oshanensis (KC460337), Buergeria buergeri (AB127977), Polypedates megacephalus (AY458598), Pelophylax nigromaculatus (AB043889), R. huanrensis (NC_028521), Hyla annectans (KM271781), O. wuchuanensis (KU680791).
Acknowledgements
The authors thank Zhibin Xiong, MAO LAN national nature reserve in Libo county, Guizhou Province, China and Zhengji Qin for their helpful assistance in the wild. The authors are grateful to Li Ding and Yanan Fan, School of Life Science, Lanzhou University, Lanzhou, China for their technical help.
Disclosure statement
The authors report no conflicts of interest. The authors are responsible for the content and alone writing of this paper.
References
- Chen Z, Zhang J, Zhai X, Zhu Y, Chen X. 2015. Complete mitochondrial genome of the green odorous frog Odorrana margaretae (Anura: Ranidae). Mitochondrial DNA. 26:487–488.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14:817–818.
- Rassmann K. 1997 . Evolutionary age of the Galápagos iguanas predates the age of the present Galápagos islands. Mol Phylogenet Evol. 7:158–172.
- Ronquist F, Teslenko M, van der Mark P, Ayres D, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Su X, Wu X-B, Yan P, Cao S-Y, Hu Y-L. 2007. Rearrangement of a mitochondrial tRNA gene of the concave-eared torrent frog, Amolops tormotus. Gene. 394:25–34.
- Zhang P, Liang D, Mao R-L, Hillis DM, Wake DB, Cannatella DC. 2013. Efficient sequencing of Anuran mtDNAs and a mitogenomic exploration of the phylogeny and evolution of frogs. Mol Biol Evol. 30:1899–1915.

