Abstract
Arini tribe with 19 genera is the most diversified tribe of neotropical parrots. Six of them are classified as macaws and nine as conures. The presence of bare facial area distinguishes macaws from conures and other members of this tribe. However, such morphological division seems to be disputable as the smallest macaw (monotypic Diopsittaca genus) turned out to be more closely related to three monotypic conures genera (Guaruba, Leptosittaca, Thectocercus) than to other macaws. We sequenced the complete mitochondrial genome of Guaruba guarouba to enrich the resource of molecular markers for examination of phylogenetic relationships between macaws and conures.
Subfamily Arinae (the New World parrots) is the most species-rich group within the order Psittaciformes (Schweizer et al. Citation2014). It is divided into four tribes (Schodde et al. Citation2013) from which the Arini tribe is the most taxon-rich. The majority of nineteen extant genera recognized within this tribe are divided into two morphologically diverse groups. Six of them (Anodorhynchus, Ara, Cyanopsitta, Diopsittaca, Orthopsittaca and Primolius) are classified as macaws (Forshaw Citation2010) based on the presence of bare facial area, which distinguishes them from other members of the tribe. Another nine genera (Aratinga, Enicognathus, Eupsittula, Guaruba, Leptosittaca, Ognorhynchus, Psittacara, Pyrrhura and Thectocercus) belong to conures (Remsen et al. Citation2016).
Recent molecular studies showed that a conure Aratinga acuticaudata should be shifted to a new genus Thectocercus acuticaudatus because it is more closely related to three monotypic genera (Diopsittaca, Guaruba and Leptosittaca) than to any member of the previously broadly defined genus Aratinga (Remsen et al. Citation2013; Urantowka et al. Citation2013). The correctness of macaws morphological classification was further undermined by the significant separation of the smallest macaw (Diopsittaca nobilis) from other members of this groups and its close relationship to conures – Guaruba, Leptosittaca and Thectocercus.
Many phylogenetic relationships within conures and macaws are still unsolved. Therefore, more molecular data are required to reconstruct precise their phylogenies. It was shown that complete mitochondrial genomes can provide useful information for evolutionary studies of many taxa (Nabholz et al. Citation2013). So far, complete mitogenomes of only three representative macaws (Ara, Orthopsittaca and Primolius) and four conures (Eupsittula, Psittacara, Pyrrhura and Thectocercus) are available (Pacheco et al. Citation2011; Urantowka et al. Citation2013; Urantowka et al. Citation2014; Urantowka Citation2016a,Citationb,Citationc; Urantowka et al. Citation2016). Therefore, we sequenced Guaruba guarouba mitogenome with the length of 17,008 bp (GeneBank accession number JQ782217) to gain appropriate molecular data for future examination of evolutionary diversification of macaws and conures.
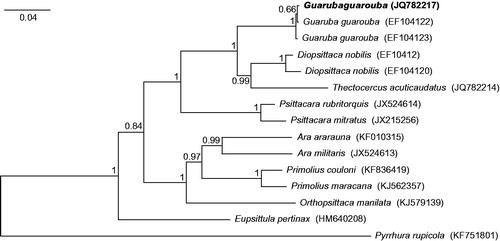
Although morphology of the analyzed specimen (Polish captive bird with CITES document no. 1281/2009 issued on 15.07.2009 in Hannover) was absolutely typical for Guaruba guarouba individuals, we aligned its control region sequence with all available other such sequences of the species as well as selected macaws and conures, to prove its species belonging. The obtained tree () revealed that the analyzed G. guarouba individual grouped significantly with two other representatives of this species. The clade Guaruba was sister to the group of Thectocercus and Diopsittaca. Three macaws (Ara, Orthopsittaca and Primolius) formed a monophyletic group but were clearly separated from the smallest macaw (Diopsittaca nobilis).
Figure 1. The phylogenetic tree obtained in MrBayes for control region sequences indicating that the studied individual (bolded) belongs to Guaruba guarouba species. The tree was generated with Bayesian method in MrBayes 3.2.5 (Ronquist et al. Citation2012) using the model GTR + I + G as suggested by jModelTest 2.1.7 (Guindon & Gascuel Citation2003; Darriba et al. Citation2012). 10,000,000 MCMC repetitions with burn-in of 25% was assumed. Tree was rooted with Pyrrhura rupicola sequence. Genbank accession numbers are shown in parenthesis. Bayesian posterior probabilities are shown at nodes.
Gene order found in G. guarouba mitogenome was the same as in Thectocercus acuticaudatus mitogenome, so far, the only fully sequenced member of the clade Guaruba/Leptosittaca/Thectocercus/Diopsittaca. The sequence identity of both genomes was 93.9% and base composition of their H-strand was nearly the same. The start and stop codons usage was also consistent for both species with the exception to the atp6 gene. In Guaruba guarouba, this gene had a truncated (TA_) stop codon, but in Thectocercus it ended with TAA codon.
Acknowledgments
We greatly thank MSc Krzysztof Grabowski for technical assistance in the laboratory.
This work is dedicated to the memory of beloved parrot named Gucio.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding
This work was supported by the National Science Centre Poland (Narodowe Centrum Nauki, Polska) under Grant no. 2015/17/B/NZ8/02402.
Additional information
Funding
References
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Forshaw JM. 2010. Parrots of the World. London: A & C Black Publishers Ltd; p. 328.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704.
- Nabholz B, Uwimana N, Lartillot N. 2013. Reconstructing the phylogenetic history of long-term effective population size and life-history traits using patterns of amino acid replacement in mitochondrial genomes of mammals and birds. Genome Biol Evol. 5:1273–1290.
- Pacheco MA, Battistuzzi FU, Lentino M, Aguilar RF, Kumar S, Escalante AA. 2011. Evolution of modern birds revealed by mitogenomics: timing the radiation and origin of major orders. Mol Biol Evol. 28:1927–1942.
- Remsen JV Jr, Areta JI, Cadena CD, Jaramillo A, Nores M, Pacheco JF, Pérez-Emán J, Robbins MB, Stiles FG, Stotz DF, Zimmer KJ. 2016. A classification of the bird species of South America. American Ornithologists' Union; [version 2016 Sept 20]. Available from: http://www.museum.lsu.edu/∼Remsen/SACCBaseline.html
- Remsen JV, Schirtzinger EE, Ferraroni A, Silveira LF, Wright TF. 2013. DNA-sequence data require revision of the parrot genus Aratinga (Aves: Psittacidae). Zootaxa. 3641:296–300.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Schodde R, Remsen JV Jr, Schirtzinger EE, Joseph L, Wright TF. 2013. Higher classification of new world parrots (Psittaciformes; Arinae), with diagnoses of tribes. Zootaxa. 3691:591–596.
- Schweizer M, Hertwig ST, Seehausen O. 2014. Diversity versus disparity and the role of ecological opportunity in a continental bird radiation. J Biogeogr. 41:1301–1312.
- Urantowka AD. 2016a. Complete mitochondrial genome of critically endangered blue-throated macaw (Ara glaucogularis): its comparison with partial mitogenome of Scarlet macaw (Ara macao). Mitochondrial DNA A DNA Mapp Seq Anal. 27:422–424.
- Urantowka AD. 2016b. Complete mitochondrial genome of Blue-headed Macaw (Primolius couloni): its comparison with mitogenome of Blue-throated Macaw (Ara glaucogularis). Mitochondrial DNA A DNA Mapp Seq Anal. 27:2106–2107.
- Urantowka AD. 2016c. Complete mitochondrial genome of Red-bellied Macaw (Orthopsittaca manilata): its comparison with mitogenome of Blue-throated macaw (Ara glaucogularis). Mitochondrial DNA A DNA Mapp Seq Anal. 27:2110–2111.
- Urantowka AD, Grabowski KA, Strzała T. 2013. Complete mitochondrial genome of Blue-crowned Parakeet (Aratinga acuticaudata)-phylogenetic position of the species among parrots group called Conures. Mitochondrial DNA. 24:336–338.
- Urantowka AD, Kroczak AM, Strzała T. 2014. Complete mitochondrial genome of endangered Socorro conure (Aratinga brevipes) - taxonomic position of the species and its relationship with green conure. Mitochondrial DNA. 25:365–367.
- Urantowka AD, Strzała T, Grabowski KA. 2016. The first complete mitochondrial genome of Pyrrhura sp.-question about conspecificity in the light of hybridization between Pyrrhura molinae and Pyrrhura rupicola species. Mitochondrial DNA A DNA Mapp Seq Anal. 27:471–473.

