Abstract
Recently resurrected Psittacara genus is one of the 19 recognized in parrot tribe Arini. The status of taxa within Psittacara remains controversial because some forms are treated as species or subspecies depending on authorities. Evolutionary history of Psittacara is also unclear because related phylogenetic clades contain taxa from distant and non-overlapping regions. However, the basal placement of Psittacara leucophthalmus with wide South American distribution suggests that other taxa with restricted range could emerge by a local split of larger population. We sequenced P. leucophthalmus mitogenome to increase the set of sequences required to determine taxonomic level and phylogeny of Psittacara taxa.
Arini tribe is the most taxon-rich among neotropical parrots (Arinae subfamily) (Schodde et al. Citation2013). The majority of Arini genera are divided into two morphologically diverse groups, macaws and conures. Among nine conure genera (Remsen et al. Citation2016), three (Eupsittula, Thectocercus, and Psittacara) were recently resurrected after the molecular revision of the previously broadly defined Aratinga genus (Remsen et al. Citation2013).
Taxonomy of Psittacara genus is still controversial. Gill and Donsker (Citation2016) recognize 11 extant Psittacara species. However, three of them (brevipes, rubritorquis, and strenuus) are treated as subspecies of holochlorus according to Dickinson and Remsen (Citation2013) and Clements et al. (Citation2016) in contrast to others (Juniper & Parr Citation1998; Forshaw Citation2010; Remsen et al. Citation2013) assuming that they have a species rank. Based on differences in plumage and habitat, Del Hoyo and Collar (Citation2014) treated frontatus (the southern P. wagleri subspecies) as a separate species. Similarly, Arndt (Citation2006) elevated the Andean P. mitratus alticola to the new species rank and described a new species P. hockingi from Peru. However, these species were not recognized by Remsen et al. (Citation2013). A museum specimen of hocking (identified as P. mitratus) seems closely related to P. wagleri but its relationship with other Psittacara was not studied molecularly.
So far, only few phylogenies for Psittacara were published. Kirchman et al. (Citation2012) using mitochondrial genes (nd2 and coxI), denied conspecificity of wagleri and mitratus species, which were suggested by Collar (Citation1997). Analyses of nd2 by Urantowka et al. (Citation2014a) showed that brevipes should be a separate species, sister to the clade of P. finschi and holochlora/rubritorquis. However, the taxonomic status of rubritorquis is still unclear, because strenuus and brewsteri (probable holochlora subspecies) were not included in the phylogenies.
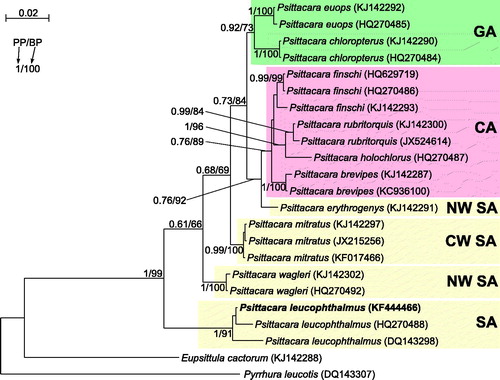
More molecular data, especially complete mitochondrial genomes (Nabholz et al. Citation2013), are required to reconstruct a precise phylogeny of Psittacara and establish a taxonomic level of its species/subspecies. Several complete mitogenomes of Psittacara taxa are already available (Urantowka et al. Citation2014a, Citation2016a, Citation2016b). To increase this set, we sequenced the mitogenome (16,966 bp, accession number KF444466) from P. leucophthalmus. Although morphology of the analyzed specimen (Polish captive bird) was absolutely typical for leucophthalmus, we proved its taxonomic affiliation in phylogenetic analyses of nd2 sequences including all available Psittacara taxa. The obtained tree () revealed that the analyzed individual groups significantly with two other representatives of its species. This species is basal to all Psittacara taxa and the next diverged lineage is P. wagleri. These positions are important to explain Psittacara diversification and migration routes, similarly to Yellow-headed Amazon parrots, for which Venezuelan Amazona barbadensis species played such pivotal role (Urantówka et al. Citation2014b). The basal placement of P. leucophthalmus and its wide distribution in South America suggests that other Psittacara species could emerge by a local split of a larger population. However, further phylogeographic history of Psittacara is complicated because subsequent clades contain parrots from distant and non-overlapping regions such as the central and northern part of South America, Central America, and Great Antilles ().
Figure 1. The phylogenetic tree obtained in MrBayes for nd2 gene indicating that the studied individual (bolded) belongs to P. leucophthalmus. The parrot is kept in aviculture and its blood sample from which DNA was isolated is available in the laboratory at the Department of Genetics in Wroclaw University of Environmental and Life Sciences under the number PL16966. Clades with taxa inhabited different geographic regions were marked by various colours/shading: CA – Central America; GA – Greater Antilles; NW SA – northwest South America; CW SA – central-western South America; SA – a vast part of South America. Migration and colonization routes of Psittacara parrots are not easy to infer because related clades include taxa, which currently have distant and restricted distributions. P. erythrogenys is the only South American species that is placed between Central American (finschi, rubritorquis, brevipes, holochlora) and Greater Antillean (euops, chloropterus) taxa. Similarly, P. wagleri with the most northern distribution in South America is placed between mitratus from the central South America, and leucophthalmus widespread in the large part of the continent. The close relationship of north-western P. erythrogenys and the Central American parrots suggests migrations through the Isthmus of Panama. However, origin of parrots from Greater Antilles remains unsolved. Values at nodes, in the order shown, indicate posterior probabilities found in MrBayes (PP) and bootstrap percentages calculated in TreeFinder (BP). In the MrBayes (Ronquist et al. Citation2012) analysis, separate mixed substitution models were assumed for three codon positions with information about heterogeneity rate across sites as proposed by PartitionFinder (Lanfear et al. Citation2012). We applied two independent runs, each using eight Markov chains. Trees were sampled every 100 generations for 10,000,000 generations. After obtaining the convergence, trees from the last 3,938,000 generations were collected to compute the posterior consensus. In the case of TreeFinder (Jobb et al. Citation2004), the separate substitution models were selected for three codon positions according to Propose Model module in this program, and 1000 replicates were assumed in the bootstrap analysis. The posterior probabilities <0.5 and bootstrap percentages <50 were omitted.
Disclosure statement
The authors report no conflicts of interest and alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Arndt T. 2006. A revision of the Aratinga mitrata complex, with the description of one new species, two new subspecies, and species-level status of Aratinga alticola. J Ornithology. 147:73–86.
- Dickinson EC, Remsen JV Jr. 2013. The Howard & Moore complete checklist of the birds of the world. 4th Edition. Vol. 1. Non-passerines. Eastbourne, UK: Aves Press.
- Clements JF, Schulenberg TS, Iliff MJ, Roberson D, Fredericks TA, Sullivan BL, Wood CL. 2016. The eBird/Clements checklist of birds of the world: v2016 [Internet]. Available from: http://www.birds.cornell.edu/clementschecklist/download/.
- Collar N, 1997. Family Psittacidae (parrots). In: J. del Hoyo, et al., editors. Handbook of the Birds of the World. Vol. 4. Sandgrouse to Cuckoos. Barcelona: Lynx Edicions; p. 280–477.
- Del Hoyo J, Collar NJ, Christie DA, Elliott A, Fishpool LDC. 2014. HBW and BirdLife international illustrated checklist of the birds of the world Volume 1: Non-passerines. Barcelona: Lynx Edicions; p. 903.
- Forshaw JM. 2010. Parrots of the world. London: A & C Black Publishers Ltd; p. 328.
- Gill F, Donsker DB, editors. 2016. IOC World Bird List (v 6.4). doi:10.14344/IOC.ML.6.4, http://www.worldbirdnames.org/.
- Jobb G, von Haeseler A, Strimmer K. 2004. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 4:18.
- Juniper T, Parr M. 1998. Parrots: a guide to parrots of the world. New Haven, London: Yale University Press.
- Kirchman JJ, Schirtzinger EE, Wright TF. 2012. Phylogenetic relationships of the extinct Carolina Parakeet (Conuropsis Carolinensis) inferred from DNA sequence data. Auk. 129:197–204.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Nabholz B, Uwimana N, Lartillot N. 2013. Reconstructing the phylogenetic history of long-term effective population size and life-history traits using patterns of amino acid replacement in mitochondrial genomes of mammals and birds. Genome Biol Evol. 5:1273–1290.
- Remsen JV Jr, Areta JI, Cadena CD, Jaramillo A, Nores M, Pacheco JF, Pérez-Emán J, Robbins MB, Stiles FG, Stotz DF, Zimmer KJ. 2016 Version [20 September 2016]. A classification of the bird species of South America [Internet]. American Ornithologists’ Union. Available: http://www.museum.lsu.edu/∼Remsen/SACCBaseline.html.
- Remsen JV, Schirtzinger EE, Ferraroni A, Silveira LF, Wright TF. 2013. DNA-sequence data require revision of the parrot genus Aratinga (Aves: Psittacidae). Zootaxa. 3641:296–300.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Schodde R, Remsen JV, Schirtzinger EE, Joseph L, Wright TF. 2013. Higher classification of New World parrots (Psittaciformes; Arinae), with diagnoses of tribes. Zootaxa. 3691:591–596.
- Urantowka AD, Kroczak AM, Strzała T. 2014a. Complete mitochondrial genome of endangered Socorro Conure (Aratinga brevipes): taxonomic position of the species and its relationship with Green Conure. Mitochondrial DNA. 25:365–367.
- Urantowka AD, Mackiewicz P, Kroczak A, Strzała T. 2016a. Complete mitochondrial genome of Red-throated Conure (Psittacara rubritorquis): its comparison with mitogenome of Socorro Conure (Psittacara brevipes). Mitochondrial DNA A DNA Mapp Seq Anal. 27:3354–3355.
- Urantówka AD, Mackiewicz P, Strzała T. 2014b. Phylogeny of Amazona barbadensis and the Yellow-headed Amazon complex (Aves: Psittacidae): a new look at South American parrot evolution. PLoS One. 9:e97228.
- Urantowka AD, Mackiewicz P, Strzała T. 2016b. Complete mitochondrial genome of Mitred Conure (Psittacara mitratus): its comparison with mitogenome of Socorro Conure (Psittacara brevipes). Mitochondrial DNA A DNA Mapp Seq Anal. 27:3363–3364.

