Abstract
In this study, we sequenced the complete mitochondrial genome (mitogenome) of the pumpkin fruit fly, Bactrocera depressa (Diptera: Tephritidae), which is an economically damaging pest of pumpkin and turban squash. The 15,832-bp-long complete mitogenome of the species consists of a typical set of genes, with an arrangement typical of insects. Of the 13 protein-coding genes (PCGs), 12 have a typical ATN start codon, whereas the COI gene begins with TCG, which has been identified as the start codon for all Bactrocera COI genes. The 1004-bp A + T-rich region of B. depressa is the third longest, after B. minax and B. scutellata, of the Bactrocera species for which the whole mitogenome has been sequenced. Phylogenetic analysis using the 13 PCGs of Bactrocera species indicated that B. depressa is a sister to the sister group containing B. tau and B. cucurbitae with the highest nodal support (Bayesian posterior probability =1.0).
The pumpkin fruit fly, B. depressa (Diptera: Tephritidae), is an economically damaging pest of pumpkin and turban squash (Han et al. Citation1994; Kim et al. Citation1999). It is distributed in the mountainous regions of Korea, Japan, and Taiwan (Han et al. Citation1994; Kim et al. Citation1999); the females lay eggs inside young pumpkins, inside which the hatched feeding larvae cause severe damage (Yoshifumi Citation1952; Kim et al. Citation1999).
In this study, we sequenced the complete mitochondrial genome (mitogenome) of B. depressa to determine the mitogenomic characteristics of this species and its phylogenetic relationships within Bactrocera. Several adults were captured from a pumpkin farm located in Gunpo City, Gyunggi-do Province, South Korea (37°19′41′′N, 126°55′55.2′′E), in September 2015. A voucher specimen was deposited in Chonnam National University, Gwangju, Korea, under the accession number CNU5743.
By using the total DNA as a template, two long overlapping fragments (COI–CytB and CytB–COI) were amplified, and subsequently 26 short overlapping fragments were amplified using the two long fragments as templates. Primers for the long and short fragments were newly designed from several existing Bactrocera mitogenome sequences (Zhang et al. Citation2014, 2015; Choudhary et al. Citation2015; Yong et al. Citation2015, Citation2016).
The B. depressa mitogenome is 15,832 bp in length and includes the typical sets of genes (2 rRNAs, 22 tRNAs, and 13 protein-coding genes [PCGs]) and a major non-coding A + T-rich region (GenBank accession number KY131831). The mitogenome size is within the range found in Bactrocera – from 15,815 bp (B. oleae; Nardi et al. Citation2003) to 16,043 bp (B. minax; Zhang et al. Citation2014). The 1004-bp A + T-rich region of B. depressa is the third longest among those of the sequenced Bactrocera, after that in B. minax (1140 bp; Zhang et al. Citation2014) and B. scutellata (1011 bp; Unpublished, GenBank accession number KP722192). In B. depressa, the A + T-rich region contains two repeat sequence composed of 33 bp that are located at the beginning and end of the A + T-rich region, respectively (15,058–15,090 and 15,780–15,812 in the genome). The gene arrangement of B. depressa is identical to that of the ancestral insect order found in the majority of insects (Boore Citation1999).
The A/T content among genes and regions varies markedly in the B. depressa mitogenome: 81.8% in the A + T-rich region, 74.5% in lrRNA, 74.5% in tRNAs, 73.1% in srRNA, and 68.8% in PCGs. 12 of the B. depressa PCGs begin with a typical ATN start codon (six with ATG, four with ATT, one with ATC, and one with ATA), whereas the COI gene begins with the atypical TCG (Serine) sequence, as has been found in all sequenced Bactrocera mitogenomes (Zhang et al. Citation2014; Choudhary et al. Citation2015; Yong et al. Citation2015, Citation2016). The B. depressa mitogenome has a total of 128 bp of intergenic spacer sequences, spread over 14 locations, ranging in size from 1 to 38 bp. Among these, three locations have spacer sequences longer than 10 bp: 38 bp between trnR and trnN, 18 bp between trnE and trnF, and 15 bp between trnS2 and ND1. These are composed of 79%, 83%, and 87% A/T nucleotides, respectively.
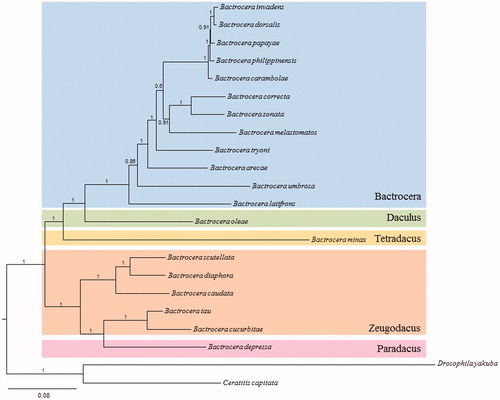
We performed a phylogenetic analysis of Bactrocera by using the 13 PCGs and two dipteran species belonging each to the same subgenus and superfamily of Bactrocera as outgroups considering a previous multigene fragment-based phylogeny of the dipteran Tribe Dacini (Krosch et al. Citation2012). The Bayesian inference method was performed using the GTR + GAMMA + I model in CIPRES Portal v. 3.1 (Miller et al. Citation2010). The results showed that B. depressa was placed as a sister to the sister group containing B. tau and B. cucurbitae with the highest nodal support (Bayesian posterior probability =1.0). Recent multigene fragment-based phylogenetic analysis of the Dacini has shown that the current subgeneric taxonomic designation of Bactrocera is not consistent with the phylogenetic clustering pattern (Krosch et al. Citation2012). Similarly, in our phylogenetic analysis, B. (Paradacus) depressa, which is classified as a member of the subgenus Paradacus, was grouped together with the sister group containing B. (Zeugodacus) tau and B. (Zeugodacus) cucurbitae, which are classified as the members of the subgenus Zeugodacus ().
Figure 1. Phylogeny of Bactrocera based on 13 protein-coding genes, constructed using the Bayesian inference method. Values at each node are percentage Bayesian posterior probabilities. The scale bar indicates the number of substitutions per site. Subgeneric names are provided on the right side of the tree. The two dipteran species D. yakuba and Ceratitis capitata were included as outgroups. GenBank accession numbers are as follows: B. zonata, KP296150 (Choudhary et al. Citation2015); B. arecae, KR233259 (Yong et al. Citation2015); B. correcta, JX456552 (Unpublished); B. dorsalis, DQ845759 (Unpublished); B. philippinensis, DQ995281 (Unpublished); B. carambolae, EF014414 (Unpublished); B. papayae, DQ917578 (Unpublished); B. tryoni, HQ130030 (Nardi et al. Citation2010); B. umbrosa, KT881558 (Yong et al. Citation2016); B. melastomatos, KT881557 (Yong et al. Citation2016); B. latifrons, KT881556 (Yong et al. Citation2016); B. invadens, KX534207 (Zhang et al. Citation2016a); B. diaphora, KT159730 (Zhang et al. Citation2016b); B. scutellata, KP722192 (Unpublished); B. tau, KP711431 (Tan et al. Citation2016); B. minax, HM776033 (Zhang et al. Citation2014); B. cucurbitae, JN635562 (Unpublished); B. caudata, KT625492 (Yong et al. Citation2016); B. oleae, AY210703 (Nardi et al. Citation2003); D. yakuba, NC001322 (Clary & Wolstenholme Citation1985); and C. capitata AJ242872 (Spanos et al. Citation2000).
Disclosure statement
A declaration of interest statement reporting no conflict has been inserted. Please confirm the statement is accurate.
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Choudhary JS, Naaz N, Prabhakar CS, Rao MS, Das B. 2015. The mitochondrial genome of the peach fruit fly, Bactrocera zonata (Saunders) (Diptera: Tephritidae): complete DNA sequence, genome organization, and phylogenetic analysis with other tephritids using next generation DNA sequencing. Gene. 569:191–202.
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22:252–271.
- Han MJ, Lee SH, Ahn SB, Choi JY, Choi KM. 1994. Distribution, damage and host plants of pumpkin fruit fly, Paradacus depressa (Shiraki). RDA J Agric Sci. 36:346–350.
- Kim TH, Kim JS, Mun JH. 1999. Distribution and bionomics of Bactrocera (Paradacus) depressa (Shiraki) in Chonbuk province. Kor J Soil Zool. 4:26–32.
- Krosch MN, Schutze MK, Armstrong KF, Graham GC, Yeates DK, Clarke AR. 2012. A molecular phylogeny for the Tribe Dacini (Diptera: Tephritidae): systematic and biogeographic implications. Mol Phylogenet Evol. 64:513–523.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proc Gateway Comput Environ Workshop (GCE); New Orleans, pp. 1–8.
- Nardi F, Carapelli A, Boore JL, Roderick GK, Dallai R, Frati F. 2010. Domestication of olive fly through a multi-regional host shift to cultivated olives: comparative dating using complete mitochondrial genomes. Mol Phylogenet Evol. 57:678–686.
- Nardi F, Carapelli A, Dallai R, Frati F. 2003. The mitochondrial genome of the olive fly Bactrocera oleae: two haplotypes from distant geographical locations. Insect Mol Biol. 12:605–611.
- Spanos L, Koutroumbas G, Kotsyfakis M, Louis C. 2000. The mitochondrial genome of the mediterranean fruit fly, Ceratitis capitata. Insect Mol Biol. 9:139–144.
- Tan M, Zhang R, Xiang C, Zhou X. 2016. The complete mitochondrial genome of the pumpkin fruit fly, Bactrocera tau (Diptera: Tephritidae). Mitochondrial DNA Part A. 27:2502–2503.
- Yong HS, Song SL, Lim PE, Chan KG, Chow WL, Eamsobhana P. 2015. Complete mitochondrial genome of Bactrocera arecae (Insecta: Tephritidae) by next-generation sequencing and molecular phylogeny of Dacini tribe. Sci Rep. 5:15155
- Yong HS, Song SL, Lim PE, Eamsobhana P, Suana IW. 2016. Complete mitochondrial genome of three Bactrocera fruit flies of subgenus Bactrocera (Diptera: Tephritidae) and their phylogenetic implications. PLoS One. 11:e0148201
- Yoshifumi T. 1952. On ecology of pumpkin fruit fly. Applied Entomol. 8:14–18.
- Zhang LJ, Jiang LH, Wei CY, Liu RS, Liu XL, Li JG, Xue HJ. 2016a. The status of Bactrocera invadens Drew, Tsuruta & White (Diptera: tephritidae) inferred from complete mitochondrial genome analysis. Mitochondrial DNA Part B. 1:680–681.
- Zhang K-J, Liu L, Rong X, Zhang G-H, Liu H, Liu Y-H. 2016b. The complete mitochondrial genome of Bactrocera diaphora (Diptera: Tephtitidae). Mitochondrial DNA Part A. 27:4314–4315.
- Zhang B, Nardi F, Hull-Sanders H, Wan X, Liu Y. 2014. The complete nucleotide sequence of the mitochondrial genome of Bactrocera minax (Diptera:Tephritidae). PLoS One. 9:e100558.

