Abstract
The complete nucleotide sequence of mitochondrial genome of the important mycoparasitic fungus Clonostachys rosea was determined using the next-generation sequencing technology. The circular molecule is 40,921 bp long with a GC content of 27.90%. Gene prediction revealed 42 genes encoding 15 conserved proteins, 25 tRNAs, the large and small ribosomal RNAs. All genes are located on the same strand. It is found to be similar to the previously sequenced mitochondrial genomes of Acremonium chrysogenum and Nectria cinnabarina. The differences lie in the copy number of trnG-UCC and locations of trnN-GUU and cox2. The phylogenetic analysis confirmed C. rosea as a sister taxon of A. chrysogenumin (Bionectriaceae). The mitochondrial genome of C. rosea will contribute to the understanding of phylogeny and evolution of Hypocreales.
Clonostachys rosea is a well-known mycoparasitic fungus used as biological control agent against several important plant pathogens, including Bipolaris sorokiana, Alternaria spp. and Fusarium spp. (Kosawang et al. Citation2014). Its nuclear genome and transcriptome analyses have been reported (Kosawang et al. Citation2014; Karlsson et al. Citation2015). The fungus is economically important and widely distributed in tropical and temperate zones. In China, it has been found from 10 provinces (Zhuang Citation2013).
The C. rosea strain 6792 was isolated from an ascoma living on bark from the Tianmu Mountain (N30°18′, E119°31′), Zhejiang Province, China. The specimen was deposited in the Mycological Herbarium, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS 183484). DNA extraction, library construction, and sequencing were performed as described in the previous studies (Wang et al. Citation2016a, Citation2016b, Citation2016c). The 125 bp pair-end reads were assembled using CLC Genomics Workbench (Version 8.0.3, CLC Bio, Aarhus, Denmark). The mitochondrial genome (mitogenome) of Trichoderma reesei (NC_003388) was served as reference to identify the assembled scaffolds belonging to mitogenome. After filtering with BLAST, only one scaffold was obtained. Manual comparison of the ends of the scaffold helped to link them into a circular molecule. This mitogenome was annotated using MFannot (Lang et al. Citation2014). Nineteen mitogenomes belonging to Hypocreales and one outgroup in Sordariales were included in the Neighbor-Joining phylogenetic analysis using MEGA6 (Tamura et al. Citation2013).
The complete sequence of C. rosea mitogenome (GenBank accession number KU668563) is 40,921 bp long with the GC content of 27.90%. It encodes 15 conserved proteins, 25 tRNAs, and the large and small ribosomal RNAs. All structural genes are located on the same strand. The tRNA genes contain codons for all 20 standard amino acids. Most amino acids are represented by only one tRNA gene; however, two trnL (trnL-UAA and trnL-UAG), two trnR (trnR-ACG and trnR-UCU), two trnS (trnS-GCU and trnS-UGA), and three trnM-CAU genes are found in this mitogenome. Eight introns are detected in five genes, i.e. rnl (1), nad5 (1), cob (1), cox1 (4), and nad1 (1).
Mitogenomes of the two members in Bionectriaceae, C. rosea and Acremonium chrysogenum (NC_023268), share the same composition and order of protein, rRNA and tRNA genes except the location of trnN-GUU and an addition of trnG-UCC for the latter. Compared with the mitogenome of Nectria cinnabarina, the representative of Nectriaceae, the quantity and order of their protein, rRNA and tRNA genes are identical except the location of cox2.
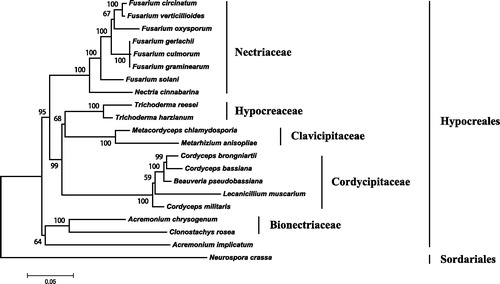
As shown in , five monophyletic clades, correlation to Bionectriaceae, Clavicipitaceae, Cordycipitaceae, Hypocreaceae, and Nectriaceae, are well resolved in the phylogenetic analysis. Clonostachys rosea and A. chrysogenum are sisters in Bionectriaceae. This family, clustered with Acremonium implicatum, islocated at the basal position of Hypocreales. This is in accordance with the combined five nuclear gene analysis (Sung et al. Citation2007). The mitogenome of C. rosea will contribute to the understanding of phylogeny and evolution of Hypocreales.
Figure 1. Phylogenetic relationship of 20 taxa of Hypocreales (Ascomycota) determined by neighbor-joining analysis based on concatenated sequences of 15 translated mitochondrial proteins. The 15 proteins included subunits of the respiratory chain complexes (cob, cox1, cox2, cox3), ATPase subunits (atp6, atp8, and atp9), NADH: quinone reductase subunits (nad1, nad2, nad3, nad4, nad4L, nad5, nad6), and ribosomal protein S3 (rps3). The concatenated sequences were aligned using MAFFT. The following 19 mitogenomes were used in this analysis: Acremonium chrysogenum (NC_023268), A. implicatum (NC_026534), Beauveria pseudobassiana (NC_022708), Cordyceps bassiana (NC_010652), C. brongniartii (NC_011194), C. militaris (NC_022834), Fusarium circinatum (NC_022681), F. culmorum (NC_026993), F. gerlachii (NC_025928), F. graminearum (NC_009493), F. oxysporum (NC_017930), F. solani (NC_016680), F. verticillioides (NC_016687), Lecanicillium muscarium (NC_004514), Metacordyceps chlamydosporia (NC_022835), Metarhizium anisopliae (NC_008068), Nectria cinnabarina (KT731105), Trichoderma harzianum (KR952346) and T. reesei (NC_003388). Neurospora crassa (NC_026614) was served as an outgroup. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the manuscript. This work was supported by the National Natural Science Foundation of China (No. 31270073 to W.-Y. Zhuang) and Beijing Natural Science Foundation of China (No. 7154224 to X.-C. Wang). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Additional information
Funding
References
- Karlsson M, Durling MB, Choi J, Kosawang C, Lackner G, Tzelepis GD, Nygren K, Dubey MK, Kamou N, Levasseur A, et al. 2015. Insights on the evolution of mycoparasitism from the genome of Clonostachys rosea. Genome Biol Evol. 7:465–480.
- Kosawang C, Karlsson M, Jensen DF, Dilokpimol A, Collinge DB. 2014. Transcriptomic profiling to identify genes involved in Fusarium mycotoxin Deoxynivalenol and Zearalenone tolerance in the mycoparasitic fungus Clonostachys rosea. BMC Genomics. 15:55.
- Lang BF, Jakubkova M, Hegedusova E, Daoud R, Forget L, Brejova B, Vinar T, Kosa P, Fricova D, Nebohacova M, et al. 2014. Massive programmed translational jumping in mitochondria. Proc Natl Acad Sci USA. 111:5926–5931.
- Sung GH, Hywel-Jones NL, Sung JM, Luangsa-Ard JJ, Shrestha B, Spatafora JW. 2007. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol. 57:5–59.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang XC, Shao J, Liu C. 2016a. The complete mitochondrial genome of the medicinal fungus Ganoderma applanatum (Polyporales, Basidiomycota). Mitochondrial DNA Part A. 27:2813–2814.
- Wang XC, Wu K, Chen H, Shao J, Zhang N, Chen X, Lan J, Liu C. 2016b. The complete mitochondrial genome of the white-rot fungus Ganoderma meredithiae (Polyporales, Basidiomycota). Mitochondrial DNA Part A. 27:4197–4198.
- Wang XC, Zeng ZQ, Zhuang WY. 2016c. The complete mitochondrial genome of the important phytopathogen Nectria cinnabarina (Hypocreales, Ascomycota). Mitochondrial DNA Part A. 27:4670–4671.
- Zhuang WY. 2013. Flora Fungorum Sinicorum vol. 47 Nectriaceae et Bionectriaceae. Beijing, China: Science Press. (In Chinese).
