Abstract
The bazaar fly, Musca sorbens (Diptera: Muscidae) Wiedemann, 1830 is a world-wide species with sanitary, medical, and veterinary importance. The complete mitochondrial genome of M. sorbens is sequenced to better understand the mitogenomic characteristics and phylogeny of this species. The circular mitogenome is 16,120 bp in length, contains 13 protein-coding genes (PCGs), 22 tRNA genes (tRNAs), two rRNA genes, and an AT-rich control region. The mitogenome comprises an A + T content of 77.4%. All PCGs start with “ATN” codons except for COI which starts with TCG, and terminate with the common stop codons TNN. A phylogenetic tree, including six Muscidae species, is reconstructed based on the whole mitogenome sequences. The interspecific distances of mitogenomes between the six Muscidae species range from 0.059 to 0.168.
Musca sorbens Wiedemann, 1830, also known as bazaar fly, belongs to Muscidae, Diptera. It is a sanitary pest that distributes all over the world, with medical and veterinary importance. Up to the present, there are 561 species belonging to this large genus (retrieved from GBIF.org Citation2018), including one of the most medically important species, Musca domestica Linnaeus, 1758. Viewing it as a potential marker like M. domestica in medical entomology, the complete mitochondrial genome sequence of M. sorbens was present here for species identification and phylogenetic analysis (GenBank accession No. MG941012).
The specimens were collected from a snake carcass in Cangyuan, Lincang, Yunnan, China (23°54′28″N, 99°13′52″E) in May 2016. All of these specimens are deposited in Department of Entomology, South China Agricultural University, Guangzhou, China (SCAU). Twenty-six overlapping fragments were amplified using total genomic DNA as templates following the study of Zhang et al. (Citation2013, Citation2015), except for the 25th primer pair was designed as 5′-AGGGTATCTAATCCTAGTT-3′ and 5′-TATAAATGGGGTATGAGCCC-3′ using available mitogenome sequences of Muscidae (Li et al. Citation2014; Lan et al. Citation2015). The amplified products were sequenced using ABI 3730xl DNA analyzer, after that assembled into a circular genome in MEGA 7.0.26 (Kumar et al. Citation2016).
The complete mitochondrial genome of M. sorbens was 16,120 bp in length, containing typical 37 genes (13 PCGs, protein-coding genes, 22 tRNA genes, and two rRNA genes) and a non-coding AT-rich control region as in other insects. Its arrangement was identical to the M. domestica mitogenome. The mitogenome of M. sorbens showed a high A + T biased (77.4%): with a base composition of A (39.1%), T (38.3%), G (9.5%), and C (13.1%). Higher A + T content of M. sorbens mtDNA was observed in the control region (86.0%). The length of tRNA genes ranged from 63 to 72 bp. All PCGs started with “ATN” codons except for COI which started with TCG, and terminate with the common stop codons TNN, which is similar to the previous results in other Diptera species (Chen et al. Citation2016; Ren et al. Citation2016; Zhu et al. Citation2016). The lrRNA and srRNA genes are 1364 and 786 bp in length, respectively.
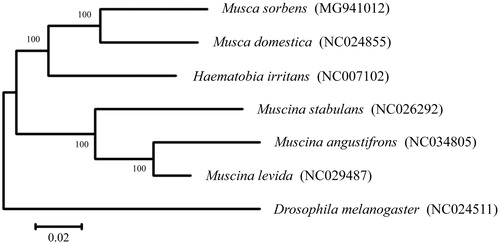
Including one outgroup taxon in Drosophilidae (Drosophila melanogaster), a phylogenetic tree of complete mitogenomes of M. sorbens and all the other available Muscidae species with Refseqs in NCBI was established using neighbour-joining (NJ) method in MEGA. In general, all the six Muscidae species were clustered but weakly supported (Bootstrap, BP lower than 50) in the NJ tree (); the outgroup taxon was clearly separated from the mitotypes of Muscidae; the three Muscina species were clustered with well supported (BP = 100). As expected, the mitogenome sequence of M. sorbens showed a close relationship (BP = 100) with the one of M. domestica. The interspecific distances of mitogenomes between the six Muscidae species ranged from 0.059 to 0.168, with an average of 0.131, and a variability of 0.032.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chen YL, Shi X, Li DY, Chen BL, Zhang P, Wu N, Xu ZX. 2016. The complete nucleotide sequence of the mitochondrial genome of Calliphora chinghaiensis (Diptera: Calliphoridae). Mitochondrial DNA B Resour. 1:397–398.
- GBIF.org 2018. GBIF Home Page. Available from: https://www.gbif.org [11 Feb. 2018].
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lan LM, Liu Y, Yan J, Lin L, Zha L. 2015. The complete mitochondrial genome of the flesh fly, Muscina stabulans (Diptera: muscidae). Mitochondrial DNA A. 27:4069–4070.
- Li XK, Wang YY, Su S, Yang D. 2014. The complete mitochondrial genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA A. 27:1435–1436.
- Ren LP, Guo QY, Yan WT, Guo YD, Ding YJ. 2016. The complete mitochondria genome of Calliphora vomitoria (Diptera: Calliphoridae). Mitochondrial DNA B Resour. 1:378–379.
- Zhang NX, Yu G, Li TJ, He QY, Zhou Y, Si FL, Ren S, Chen B. 2015. The complete mitochondrial genome of Delia antiqua and its implications in dipteran phylogenetics. PLoS One. 10:e0139736.
- Zhang NX, Zhang YJ, Yu G, Chen B. 2013. Structure characteristics of the mitochondrial genomes of Diptera and design and application of universal primers for their sequencing. Acta Entomol Sin. 56:398–407.
- Zhu ZY, Liao HD, Ling J, Guo YD, Cai JF, Ding YJ. 2016. The complete mitochondria genome of Aldrichina grahami (Diptera: Calliphoridae). Mitochondrial DNA B Resour. 1:107–109.