Abstract
Rockfishes of the genus Sebastes rank among the longest-lived vertebrate animals. In order to facilitate comparative genomic research in animal longevity, the complete mitochondrial genome sequences are presented for Sebastes aleutianus, Sebastes minor, Sebastes nigrocinctus, Sebastes rubrivinctus, and Sebastes steindachneri.
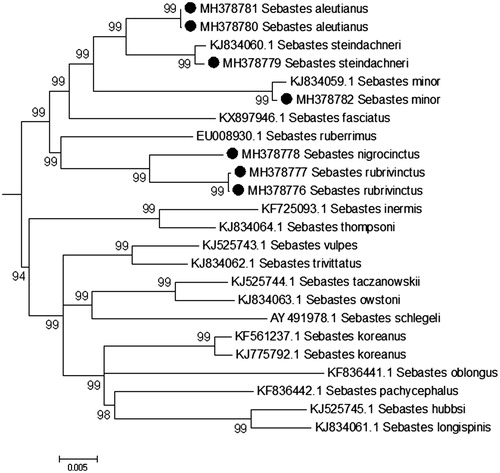
Within the Perciformes genus Sebastes (Rockfishes) rank among the most long-lived bony vertebrates, with age estimates of multiple species exceeding 150 years (Cailliet et al. Citation2001). Rockfishes have been notable subjects of the comparative biology of aging, and the mitochondrial genome has provided insight into the evolution of lifespan within the genus (Heras et al. Citation2011, Citation2015; Hua et al. Citation2015). The Free Radical Theory of Aging posits the electron transport chain as a fundamental determinant of aging and lifespan, via the production and escape of free radicals from the mitochondrion (Harman Citation1992). We sequenced the complete mitochondrial genomes of five rockfish species to facilitate comparative functional genomics research relevant to the study of aging. This is the first report of complete mitochondrial sequences for Sebastes aleutianus (n = 2), Sebastes nigrocinctus (n = 1), Sebastes rubrivinctus (n = 2), and Sebastes minor (n = 1). We also report a new mtDNA sequence of Sebastes steindachneri (n = 1), corroborating results of Jang, Lee, et al. (Citation2016). Specimens were collected via trawl from the eastern North Pacific Ocean. Specimens were identified and fin-clipped by J.R. Hyde, A.J. Gharrett, and Y. Kai. Morphological vouchers were not retained, but nuclear genome sequences are available for species validation from the Nucleotide database at ncbi.nlm.nih.gov. DNA was extracted using the Qiaxcel DNeasy protocol (Qiagen). Library preparation followed TruSeq protocol for illumina paired-end high-throughput DNA sequencing. Data were generated using an Illumina HiSeq 2000 at the University of Southern California. Paired-end mtDNA sequence reads were selectively assembled to build the complete mitochondrial genome using the iterative mapping approach of MitoBim 1.7 (Hahn et al. Citation2013). The genome of Sebastes koreanus (accession KJ775792.1) was used as a reference sequence. Genome assemblies from each of the paired output files were aligned to each other using the MAFFT online alignment tool (Katoh et al. Citation2017). Consensus sequences for each sample revealed a standard gene order and GC% content for teleost fishes. Genome annotation was conducted using the online MitoFish MitoAnnotator tool (Iwasaki et al. Citation2013). The Cytochrome Oxidase 1 gene was used to corroborate species identifications made in the field. For each species, a BLAST search of the CO1 sequence against the NCBI Nucleotide database revealed at least 99% identity to accessions of the same species name. Variability among protein-coding and tRNA genes revealed few insertion-deletions and modest levels of substitutional variation. Conversely, the 3′ region of the mtDNA control region exhibited relatively high levels of sequence divergence and copy number variation, consistent with previous observations of rockfishes (Bentzen et al. Citation1998; Higuchi and Kato Citation2002; Zhang et al. Citation2013). Specifically, a 200–300 bp sequence repeat was detected in the 3′ region of the mtDNA control region. This large repeat sequence varied among species and among individuals within species. The algorithm used for genome assembly did not allow precise quantitation of copy number. Seventeen Sebastes mitochondrial genomes were obtained from ncbi.nlm.nih.gov for comparative analysis. A minimum evolution phylogenetic analysis, generated with MEGA7 reveals strong support for sister-taxon relationships for species sequenced herein () (Kumar et al. Citation2016). In this taxon-limited analysis, Sebastes aleutianus was sister to S. steindachneri, and this clade was sister to S. minor. S. nigrocinctus was sister to Sebastes rubrivinctus. All other relationships were consistent with results of previous studies (Jang, Kim, Kim Citation2015; Jang, Kim, Oh, et al. Citation2015; Jang, Lee, et al. Citation2016; Jang, Oh, Park, et al. Citation2016; Jang, Oh, Lee, et al. Citation2016; Jang, Park, et al. Citation2016).
Figure 1. Sebastes phylogeny inferred using the Minimum Evolution (ME) method. The optimal tree with the sum of branch length = 1.07547521 is shown. Evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. The ME tree was searched using the Close-Neighbor-Interchange (CNI) algorithm at a search level of 1. All positions containing gaps and missing data were eliminated. There were a total of 13,452 positions in the final dataset. Outgroups are not shown.
Disclosure statement
No potential conflict of interest was reported by the authors.
Reference
- Bentzen P, Wright JM, Bryden LT, Sargent M, Zwanenburg KCT. 1998. Tandem repeat polymorphism and heteroplasmy in the mitochondrial control region of redfishes (Sebastes: Scorpaenidae). J Hered. 89:1–7.
- Cailliet GM, Andrews AH, Burton EJ, Watters DL, Kline DE, Ferry-Graham LA. 2001. Age determination and validation studies of marine fishes: do deep-dwellers live longer? Exp Gerontol. 36:739–764.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129.
- Harman D. 1992. Free radical theory of aging. Mutat Res. 275:257–266.
- Heras J, Koop BF, Aguilar A. 2011. A transcriptomic scan for positively selected genes in two closely related marine fishes: Sebastes caurinus and S. rastrelliger. Mar Genomics. 4:93–98.
- Heras J, McClintock K, Sunagawa S, Aguilar A. 2015. Gonadal transcriptomics elucidate patterns of adaptive evolution within marine rockfishes (Sebastes). BMC Genomics. 16:656.
- Higuchi M, Kato K. 2002. Sequence variability in the mitochondrial DNA control region of five Sebastes species. Fisheries Sci. 68:643–650.
- Hua X, Cowman P, Warren D, Bromham L. 2015. Longevity is linked to mitochondrial mutation rates in rockfish: a test using poisson regression. Mol Biol Evol. 32:2633–2645.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, Nishida M. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Jang Y-S, Kim K-Y, Kim S. 2015. The complete mitochondrial genome of the Hwanghae Rockfish Sebastes koreanus (Scorpaenidae, Scorpaeniformes). Mitochondrial DNA. 26:834–835.
- Jang Y-S, Kim K-Y, Oh S-Y, Choi H-J, Myoung J-G, Kim S. 2015. The complete mitochondrial genome of the dark-banded rockfish Sebastes inermis (Scorpaenidae, Scorpaeniformes). Mitochondrial DNA. 26:895–896.
- Jang Y-S, Lee EK, Park J-H, Kim K-Y, Kim S. 2016. Complete mitochondrial genome of Sebastes steindachneri (Scorpaenidae: Scorpaeniformes) from the East Sea, Korea. Mitochondrial DNA Part A. 27:4041–4043.
- Jang Y-S, Oh S-Y, Lee EK, Myoung J-G, Park J-H, Kim K-Y, Kim S. 2016. Complete mitochondrial genome of Sebastes vulpes (Scorpaenidae, Scorpaeniformes) from the East Sea, Korea. Mitochondrial DNA Part A. 27:1239–1240.
- Jang Y-S, Oh S-Y, Park K-J, Kim K-Y, Kim S. 2016. The complete mitochondrial genome of the oblong rockfish Sebastes oblongus (Scorpaenidae, Scorpaeniformes). Mitochondrial DNA Part A. 27:65–66.
- Jang Y-S, Park K-J, Kim K-Y, Kim S. 2016. The complete mitochondrial genome of Sebastes pachycephalus (Scorpaenidae, Scorpaeniformes) from the East Sea, Korea. Mitochondrial DNA Part A. 27:69–70.
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. https://doi.org/10.1093/bib/bbx108
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Zhang H, Zhang Y, Zhang X, Song N, Gao T. 2013. Special structure of mitochondrial DNA control region and phylogenetic relationship among individuals of the black rockfish, Sebastes schlegelii. Mitochondrial DNA. 24:151–157.

