Abstract
We present the complete mitochondrial genome of Pomacea maculate in this study. The mitochondrial genome is 15,512 bp in length, containing 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes. Overall nucleotide compositions of the light strand are 41.13% of A, 30.81% of T, 15.25% of C and 12.81% of G. Its gene arrangement and distribution are different from the typical vertebrates. The absence of D-loop is consistent with the Gastropoda, but at least one lengthy non-coding region is essential regulatory element for the initiation of transcription and replication. Phylogenetic tree is constructed by the maximum-likelihood method based on the complete mitochondrial genomes of 15 species of Caenogastropoda, using Helix aspersa as outgroup to assess their actual phylogenetic relationship and evolution. The result provides fundamental data for resolving phylogenetic and genetic problems related to effective management strategies.
Pomacea maculata (formerly P. insularum) (Perry, 1810), was introduced to Asia from Brazil and Argentina independently (Hayes et al. Citation2008). Subsequently, Lv et al. (Citation2013) reported that P. maculata, was established in China. Pomacea maculata is one of two most common and highly invasive apple snail species as P. canaliculata (Lamarck, 1822) currently (Hayes et al. Citation2012), which strips vegetation, reproduces at tremendous rates, and have reduced rice production and caused ecosystem issues in Asia (Pimentel et al. Citation2005; Zedler and Kercher Citation2005; Burlakova et al. Citation2009). Some studies confirmed a much lower haplotype diversity of P. maculata in populations of China than that in their native countries Argentina and Brazil with a statistic ratio of 3:34 (Hayes et al. Citation2008; Lv et al. Citation2013; Yang et al. Citation2018). Furthermore, just a single lineage of P. maculata from Brazil was introduced into and established in China.
We sequenced its complete mitogenome to analyze phylogenetic relationship and evolutionary history for broadening the understanding of their population diversity, invasion processes and implementing effective management strategies. The specimen was sampled from Ningxi Teaching and Research Farm of South China Agricultural University in Guangzhou (E 113°29′, N 23°5′), and stored in the specimen museum of SCAU (accession number: 201502118).
The complete mitochondrial genome of P. maculate (Genbank accession number KY008699) is 15,512 bp in length, containing 13 protein-coding genes, 2 ribosomal RNA genes (rrns and rrnl), 22 transfer RNA genes (tRNA), which are encoded on the heavy strand except 8 tRNA genes (Met, Tyr, Cys, Trp, Gln, Gly, Glu and Thr) on the light strand. 22 tRNA genes vary from 64 to 75 bp in length, and all fold into the typical cloverleaf secondary structure. Among 13 protein-coding genes (total 11,238 bp) encoding 3,733 amino acids, the maximum is ND5 with 1,710 bp, and the minimum is ATP8 with only 159 bp. Rrns and rrnl genes are 934 and 1331 bp, respectively, located between the tRNAGlu and tRNALeu genes and separated by the tRNAVal gene. Overall nucleotide compositions of the light strand in descending order are 41.13% of A, 30.81% of T, 15.25% of C and 12.81% of G. Gene arrangement and distribution are different from the typical vertebrates (Yang, Sun, Zhao, Chen, et al. Citation2016; Yang, Sun, Zhao, Yang, et al. Citation2016; Yang, Xie, et al. Citation2016; Yang, Zhao, Sun, Chen, et al. Citation2016; Yang, Zhao, Sun, Xie, et al. Citation2016; Yang, Zhao, Sun, Yang, et al. Citation2016; Yang, Zhao, Sun, Zhang, et al. Citation2016; Yang, Zhao, Xie, et al. Citation2016), and similar to the Mollusca (Yang, Zhang, Deng, et al. Citation2016; Yang, Zhang, Guo, et al. Citation2016; Yang, Zhang, Luo, et al. Citation2016; Guo et al. Citation2017). The absence of D-loop is consistent with the Gastropoda (Yang, Zhang, Deng, et al. Citation2016; Yang, Zhang, Guo, et al. Citation2016; Yang, Zhang, Luo, et al. Citation2016; Guo et al. Citation2017), but at least one lengthy non-coding region is essential regulatory element for the initiation of transcription and replication (Wolstenholme Citation1992). There are 24 intergenic spacers (total 547 bp) varying from 3 to 141 bp in length, the largest of which is 141 bp between tRNA-Phe and NC III gene, and 2 gene overlaps (total 13 bp).
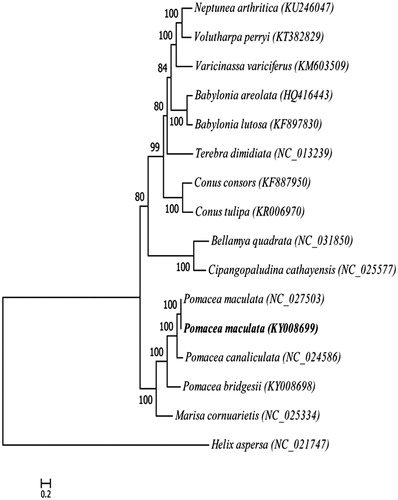
Phylogenetic tree is constructed by the maximum-likelihood method based on the complete mitogenomes of 15 species of Caenogastropoda, using Helix aspersa as outgroup to assess their actual phylogenetic relationship and evolution ().
Figure 1. Phylogenetic tree generated by the maximum-likelihood method based on the complete mitochondrial genomes of 15 species of Caenogastropoda, using Helix aspersa as outgroup. The published sequences in GenBank adopted are Neptunea arthritica (KU246047), Volutharpa perryi (KT382829), Varicinassa variciferus (KM603509), Babylonia areolata (HQ416443), Babylonia lutosa (KF897830), Terebra dimidiate (NC_013239), Conus consors (KF887950), Conus tulipa (KR006970), Bellamya quadrata (NC_031850), Cipangopaludina cathayensis (NC_025577), Pomacea maculate (NC_027503), Pomacea maculate (KY008699), Pomacea canaliculata (NC_024586), Pomacea bridgesii (KY008698), Marisa cornuarietis (NC_025334), Helix aspersa (NC_021747).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Burlakova LE, Karatayev AY, Padilla DK, Cartwright LD, Hollas DN. 2009. Wetland restoration and invasive species: apple snail (Pomacea insularum) feeding on native and invasive aquatic plants. Restor Ecol. 17(3):433–440.
- Guo J, Yang HR, Zhang CX, Xue HY, Xia Y, Zhang JE. 2017. Complete mitochondrial genome of the apple snail Pomacea diffusa (Gastropoda, Ampullariidae) with phylogenetic consideration. Mitochondrial DNA B. 2(2):865–867.
- Hayes KA, Cowie RH, Thiengo SC, Strong EE. 2012. Comparing apples with apples: clarifying the identities of two highly invasive neotropical Ampullariidae (Caenogastropoda). Zool J Linn Soc-Lond. 166(4):723–753.
- Hayes KA, Joshi RC, Thiengo SC, Cowie RH. 2008. Out of South America: multiple origins of non-native apple snails in Asia. Divers Distrib. 14(4):701–712.
- Lv S, Zhang Y, Liu HX, Hu L, Liu Q, Wei FR, Guo YH, Steinmann P, Hu W, Zhou XN, Utzinger J 2013. Phylogenetic evidence for multiple and secondary introductions of invasive snails: Pomacea species in the People's Republic of China. Diversity Distrib. 19(2):147–156.
- Pimentel D, Zuniga R, Morrison D. 2005. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol Econ. 52(3):273–288.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.
- Yang HR, Sun JJ, Zhao HH, Chen YB, Yang ZT, Li GF, Liu L. 2016. The complete mitochondrial genome of the Clarias fuscus (Siluriformes, Clariidae). Mitochondrial DNA A. 27(2):1255–1256.
- Yang HR, Sun JJ, Zhao HH, Yang ZT, Xiao SB, Li GF, Liu L. 2016. The complete mitochondrial genome of Nibea coibor (Perciformes, Sciaenidae). Mitochondrial DNA A. 27(2):1530–1522.
- Yang Y, Xie Z, Peng C, Wang J, Li S, Zhang Y, Zhang H, Lin H. 2016. The complete mitochondrial genome of the orange-spotted grouper Epinephelus coioides (Perciformes, Serranidae). Mitochondrial DNA A. 27(3):1674–1676.
- Yang HR, Zhang JE, Deng ZX, Luo H, Guo J, He SM, Luo MZ, Zhao BL. 2016. The complete mitochondrial genome of the golden apple snail Pomacea canaliculata (Gastropoda: Ampullariidae). Mitochondrial DNA B. 1(1):45–47.
- Yang HR, Zhang JE, Guo J, Deng ZX, Luo H, Luo MZ, Zhao BL. 2016. The complete mitochondrial genome of the giant African snail Achatina fulica (Mollusca: Achatinidae). Mitochondrial DNA A. 27(3):1622–1624.
- Yang HR, Zhang JE, Luo H, Luo MZ, Guo J, Deng ZX, Zhao BL. 2016. The complete mitochondrial genome of the mudsnail Cipangopaludina cathayensis (Gastropoda: Viviparidae). Mitochondrial DNA A. 27(3):1892–1894.
- Yang HR, Zhao HH, Sun JJ, Chen YB, Liu LJ, Zhang Y, Liu L. 2016. The complete mitochondrial genome of the Hemibarbus medius (Cypriniformes, Cyprinidae). Mitochondrial DNA A. 27(2):1070–1072.
- Yang HR, Zhao HH, Sun JJ, Xie ZZ, Yang ZT, Liu L. 2016. The complete mitochondrial genome of the Culter recurviceps (Teleostei, Cyprinidae). Mitochondrial DNA A. 27(1):762–763.
- Yang HR, Zhao HH, Sun JJ, Yang ZT, Xiao SB, Li GF, Liu L. 2016. The complete mitochondrial genome of the amoy croaker Argyrosomus amoyensis (Perciformes, Sciaenidae). Mitochondrial DNA A. 27(2):1530–1532.
- Yang HR, Zhao HH, Sun JJ, Zhang Y, Yang ZT, Liu L. 2016. The complete mitochondrial genome of the Hemibagrus wyckioides (Siluriformes, Bagridae). Mitochondrial DNA A. 27(1):766–768.
- Yang HR, Zhao HH, Xie ZZ, Sun JJ, Yang ZT, Liu L. 2016. The complete mitochondrial genome of the Hemibagrus guttatus (Teleostei, Bagridae). Mitochondrial DNA A. 27(1):679–681.
- Yang QQ, Liu SW, He C, Yu XP. 2018. Distribution and the origin of invasive apple snails, Pomacea canaliculata and P. maculata (Gastropoda: Ampullariidae) in China. Sci Rep. 8:1185.
- Zedler JB, Kercher S. 2005. Wetland resources: status, trends, ecosystem services, and restorability. Annu Rev. Environ. Resour. 30:39–74.

