Abstract
Genomic analysis of the kelp fly Fucellia costalis (Family Anthomyiidae) from Asilomar beach, Pacific Grove, California, resulted in the assembly of its complete mitogenome. The mitogenome is 16,175 bp in length, A + T biased (78.1%), contains 37 genes, a large 1356 bp control region, and has a high-level of gene synteny to other Muscoidea. Phylogenetic analysis of F. costalis fully resolves it in a clade with two members of the Anthomyiidae, Delia platura (8.2% pairwise genetic distance) and D. antiqua (9.2% distance), in a sister position to Scathophaga stercoraria (7.8% distance) from the Scathophagidae. The phylogenetic position and genetic distances of F. costalis support the continued recognition of the subfamily Fucellinae. This identification and data extends the distributional range of F. costalis northward from California to Vancouver Island, Canada.
The Dipteran family Anthomyiidae consists of approximately 1100 species that occur worldwide and inhabit a diversity of habitats including seashores, gardens, woods, open fields, and alpine meadows (Huckett Citation1971; Couri and Rodrigues-Júnior Citation2012). A single complete mitogenome was published for the family of Delia antiqua, subfamily Anthomyiinae (Zhang et al. Citation2015). Here, we describe the mitogenome of a kelp fly collected from Asilomar beach, Pacific Grove, California (36°37'43.2"N, −121°55'17.5"W), Fucellia costalis, an abundant swarming Dipteran and the largest of 26 species in Fucellia, subfamily Fucellinae (Aldrich Citation1918; Cole Citation1969).
DNA was extracted from F. costalis (Specimen Voucher- Hughey, 17 September 2016, J. Gordon Edwards Musuem, San Jose State University) following the protocol of Lindstrom et al. (Citation2011). The 76 and 150 bp paired-end Illumina library constructions and sequencing were performed by myGenomics, LLC (Alpharetta, GA). The mitogenome was assembled using default de novo settings in MEGAHIT (Li et al. Citation2015). The single gap that resulted was closed using standard PCR and Sanger sequencing methods (Lindstrom et al. Citation2011) with the following primers: F/d-loop 5′-GTTCATAACTAATAAGATTAATAGATATC-3′ and R/trnaMet 5′-GATTAGAACCTTTATAAATGGGG-3′. The genes were annotated manually by aligning the F. costalis mitogenome to Scathophaga stercoraria (GenBank accession KM200724) in Geneious R11 (Biomatters Limited, Auckland, New Zealand). Alignment of F. costalis to other complete Muscoidea mitogenomes was accomplished with MAFFT (Katoh and Standley Citation2013). The Maximum-likelihood analysis was executed using RaxML in Trex-online (Boc et al. Citation2012) with the GTR + gamma model and 1000 bootstraps. The tree was visualized with TreeDyn 198.3 at Phylogeny.fr (Dereeper et al. Citation2008).
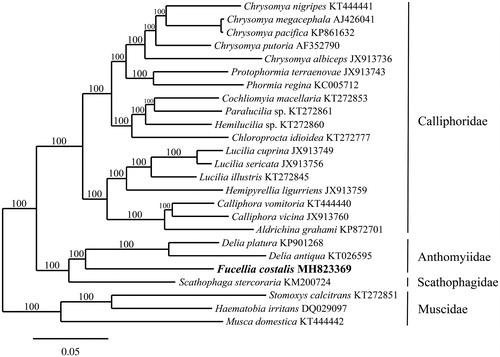
The mitogenome of F. costalis (GenBank accession number MH823369) is 16,175 bp in length and A + T rich with base a composition of 40.0% A, 38.1% T, 9.4% G, and 12.5% C. It is similar in length, content, and gene organization to other Muscoidea (Ding et al. Citation2015; Zhang et al. Citation2015; Li et al. Citation2016). The mitogenome contains 22 tRNA (trnL and trnS are duplicated), two rRNA (rrnL, rrnS), 13 genes involved in electron transport and oxidative phosphorylation, and a control region that is 1356 bp in length. Six of the coding genes are initiated with the ATG codon (ATP6, COII, COIII, CytB, ND4, ND4L), four with ATT (ATP8, ND2, ND5, ND6), two with ATA (ND1, ND3), and one with ATC (COI). Nine of the coding genes terminate with the stop codon TAA, however COI, COII, ND4, and ND5 terminate with the incomplete codon T. Eight tRNAs, ND1, ND4, ND4L, ND5, and the two rRNAs encode on the light-strand, with the remaining genes encoding on the heavy-strand. Phylogenetic analysis of the F. costalis mitogenome resolves it in a fully supported clade with Delia platura (8.2% pairwise genetic distance) and D. antiqua (9.2% distance) (subfamily Anthomyiidae), sister in position to Scathophaga stercoraria (7.8% distance) (). A BLAST search of the F. costalis mitogenome found a single genetic match of an unidentified Anthomyiid from Vancouver Island (COI GenBank accession JF867328), extending the known distribution of F. costalis from California to Canada.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Aldrich JM. 1918. The kelp-flies of North America (genus Fucellia family Anthomyidae). Proc Calif Acad Sci, Ser 4. 8:157–179.
- Boc A, Diallo AB, Makarenkov V. 2012. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 40:W573–W579.
- Cole FR. 1969. The flies of Western North America. Berkeley, CA: University of California Press.
- Couri MS, Rodrigues-Júnior FA. 2012. First record of Anthomyiidae (Diptera) from New Caledonia with key to Australasian and Oceanian species of Anthomyia Meigen. Rev Bras Entomol. 56:183–185.
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. 2008. Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469.
- Ding S, Li X, Wang N, Cameron SL, Mao M, Wang Y, Xi Y, Yang D. 2015. The phylogeny and evolutionary timescale of Muscoidea (Diptera: Brachycera: Calyptratae) inferred from mitochondrial genomes. PLoS ONE. 10:e0134170.
- Huckett HC. 1971. The Anthomyiidae of California exclusive of the subfamily Scatophaginae (Diptera). Bull Calif Insect Survey. 12:1–121.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 31:1674–1676.
- Li XK, Wang YY, Su S, Yang D. 2016. The complete mitochondrial genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA. 27:1435–1436.
- Lindstrom SC, Hughey JR, Martone PT. 2011. New, resurrected and redefined species of Mastocarpus (Phyllophoraceae, Rhodophyta) from the northeast Pacific. Phycologia. 50:661–683.
- Zhang N-X, Yu G, Li T-J, He Q-Y, Zhou Y, Si F-L, Ren S, Chen B. 2015. The Complete mitochondrial genome of Delia antiqua and its implications in Dipteran phylogenetics. PLoS One. 10:e0139736.