Abstract
The crane fly Tipula (Formotipula) melanomera gracilispina belongs to the Tipulidae. The mitogenome of T. (F.) melanomera gracilispina was sequenced, the new representative of the mitogenome of the family. The nearly complete mitogenome is 14,575 bp totally, consisting of 13 protein-coding genes, two rRNAs and 22 transfer RNAs. All genes have the similar locations and strands with that of other published species of Tipuloidea. The nucleotide composition biases toward A and T, which together made up 76.3% of the entirety. Bayesian inference analysis strongly supported the monophyly of Tipuloidea. It suggested that monophyletic Tipulidae was assigned to the sister to the monophyletic Cylindrotomidae and the Limoniidae is the sister group to the clade of Tipulidae + Cylindrotomidae.
Introduction
With more than 15,000 described species in over 700 genera and subgenera, Tipuloidea is one of the most diverse groups in Diptera (Oosterbroek Citation2018), and this diversity reflects the extensive morphological and ecological adaptations of its members.
The specimens of T. (F.) melanomera gracilispina used for this study were collected in Sam Nuea City of Houaphanh Province of Laos by Liang Wang (20°24′N, 104°2′E), and then identified by Bing Zhang. Specimens are deposited in the Entomological Museum of China Agricultural University, Beijing. The genomic DNA was extracted from the whole body of the specimen using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) and stored at −20 °C until needed. The mitogenome was amplified and sequenced as described in our previous study (Wang et al. Citation2016). The nearly complete mitogenome of T. (F.) melanomera gracilispina is 14,575 bp. It encoded 13 PCGs, 22 tRNA genes and two rRNA genes and the control region could not be sequenced in this study, and were similar with related reports before (Li et al. Citation2016, Citation2017). All genes have the similar locations and strands with that of other published Tipuloidea species. The nucleotide composition of the mitogenome was biased toward A and T, with 76.3% of A + T content (A = 39.0%, T = 37.3%, C = 14.7%, G = 9.1%). The A + T content of PCGs, tRNAs, and rRNAs is 75.6%, 77.8%, and 79.2% respectively. The total length of all 13 PCGs of T. (F.) melanomera gracilispina is 11,209 bp. Three PCGs (NAD2, NAD3 and NAD6) initiated with ATT codons, and six PCGs (COII, COIII, ATP6, NAD4L, NAD4 and CYTB) initiated with ATG codons, ATP8 initiated with ATC as a start codon, NAD1 initiated with ATA as a start codon, CO1 and NAD5 initiated with TCG and GTG as a start codon respectively. Six PCGs used the typical termination codons TAA, three PCGs (CO2, ATP6 and NAD5) used T, three PCGs (NAD1, NAD3 and CYTB) used TAG, and one PCG (NAD4) used TA in T. (F.) melanomera gracilispina.
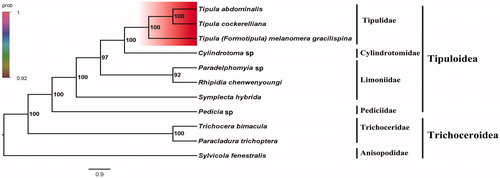
Phylogenetic analysis was performed based on the nucleotide sequences of 13 PCGs from 11 Diptera species. Bayesian (BI) analysis generated the phylogenetic tree topologies based on the PCGs matrices (). According to the phylogenetic result, it showed that monophyletic Tipuloidea was assigned to be the sister group to the clade of Trichoceroidea with Trichoceridae in this text. The monophyletic Tipulidae was the sister group to the monophyletic Cylindrotomidae. For the phylogeny of Tipuloidea, the clade consisting of Tipulidae + Cylindrotomidae is the sister group to the clade of Limoniidae. The phylogenetic relationship inferred from the phylogenetic result in this text is very clear: Trichoceroidea + (Pediciidae + (Limoniidae + (Tipulidae + Cylindrotomidae))). The relationship of the sister group within Tipuloidea was also supported by the previous study (Petersen et al. Citation2010). The mitogenome of T. (F.) melanomera gracilispina could provide the important information for the further studies of Tipuloidea phylogeny.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Li X, Ding S, Hou P, Liu X, Zhang C, Yang D. 2017. Mitochondrial genome analysis of Ectophasia rotundiventris (Diptera: Tachinidae). Mitochondrial DNA Part B. 2:457–458.
- Li X, Wang Y, Su S, Yang D. 2016. The complete mitochondrial genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1435–1436.
- Oosterbroek P. 2018. Catalogue of the Craneflies of the World [WWW document]. URL http://ip30.eti.uva.nl/ccw/ [accessed on 6 September 2018].
- Petersen MJ, Bertone MA, Wiegmann BM, Courtney GW. 2010. Phylogenetic synthesis of morphological and molecular data reveals new insights into the higher-level classification of Tipuloidea (Diptera). System Entomol. 35:526–545.
- Wang K, Li X, Ding S, Wang N, Mao M, Wang M, Yang D. 2016. The complete mitochondrial genome of the Atylotus miser (Diptera: Tabanomorpha: Tabanidae), with mitochondrial genome phylogeny of lower Brachycera (Orthorrhapha). Gene. 586:184–196.