Abstract
Xenocypris yunnanensis is an endemic species subsisted in Dianchi Lake in China and has been listed as one of the most dangerous species due to the invasion of alien species. In the present study, we determined the first complete mitochondrial genome of X. Yunnanensis, This genome was 16,630 bp in size and contained 37 genes (13 protein-coding genes, two ribosomal RNA, 22 transfer RNA genes and one control region). The overall base composition of the heavy strand is that the 31.25% for A, the 27.20% for C, the 16.15% for G and the 25.40% for T. In this study, the complete mitochondrial genome sequencing for X. yunnanensis was determined for the first time. The result of phylogenetic analysis indicated is that X. yunnanensis is clustered in genus Xenocypris as expected. Also, The mitochondrial genome sequencing of X. yunnanensis provided important molecular data for conservation genetic studies.
Introduction
Xenocypris yunnanensis belongs to the order Cypriniformes, Cyprinidae family, and genus Xenocyprinae. Xenocypris yunnanensis is an endemic species in Dianchi Lake, Kunming, Yunnan, China which is (24°40′–25°02′ N, 102°36′–102°47′ E,1,885 m above sea level and covered 306.3 km2 (Liu et al. Citation2009). It has not been seen by frequent aquatic biodiversity surveys (GEF/The World Bank) in the lake and it is considered endangered now. Their greatest threats came through introduction of commercial fishes, such as Hemisalanx prognathous into Dianchi Lake X. yunnanensis has already been listed as the most endangered species by the red list of IUCN (2008), as endangered in China Red Data Book of Endangered Animals (Wang and Xie Citation2004) and China Species Red List, Vol. 1 Red List (Sung et al. Citation1998).
In this work, a matured X. yunnanensis collected from Dianchi Lake during catching season in 2016 by accident was used for the complete mitochondrial DNA sequencing. The total genomic DNA was extracted from alcohol-preserved fin using the traditional phenol-chloroform method (Taggart et al. Citation1992). Total genomic DNA was extracted from tail tip using the Ezup pillar genomic DNA extraction kit (Sangon Biotech, Shanghai, China). A total of 13 pairs of primers, designed based on the mitochondrial genome of Xenocypris davidi (KF039718.1) (Liu Citation2014), were used to amplify the complete mitochondrial genome. The PCR products were sequenced using the Sanger method and further assembled into a sequence with CAP3 (Huang and Madan Citation1999).
The complete mitogenome sequence of X. yunnanensis is16,630 bp in length (GenBank accession number KY993905) and consists of 13 protein-coding genes, two ribosomal RNA, 22 transfer RNA genes and one control region. Among these genes, ND6, tRNA-GAA, tRNA-CCA, tRNA-CAA, tRNA-GCA, tRNA-CGA, tRNA-UAC, and tRNA-UCA are located on the light strand (L-strand), while all the remaining genes on the heavy strand (H-strand). The overall base composition of the heavy strand is 31.25% for A, 27.20% for C, 16.15% for G and the 25.40% for T. The percentage of GC content is 43.35% and AT content is 56.65%.
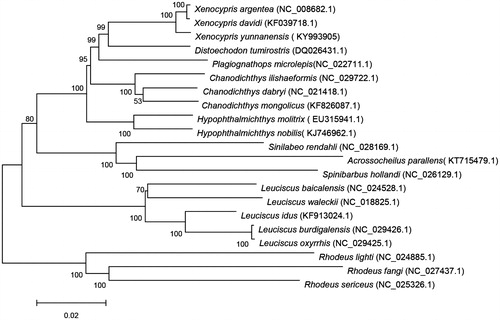
We constructed a phylogenetic tree including X. yunnanensis and the other 20 fish. Phylogenetic relationships obtained with the maximum likelihood approach were identical to those of the Bayesian analysis (Posada and Buckley Citation2004; Wang et al. Citation2015). As shown in the phylogenetic tree (), X. yunnanensis sequence was clustered in genus Xenocyprinae, including Xenocypris argentea, Xenocypris davidi, Distoechodon tumirostris and Plagiognathops microlepis. Phylogenetic analysis revealed that X. yunnanensis is closely related to X. argentea and X. davidi.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868–877.
- Liu SW, Chen XY, Yang JX. 2009. Threatened fishes of the world: Xenocypris yunnanensis (Nichols, 1925) (Cypriniformes: Cyprinidae). Environ Biol Fishes. 85:361–362.
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 53:793–808.
- Taggart JB, Hynes RA, Prodouhl PA, Ferguson A. 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol. 40:963–965.
- Liu Y. 2014. The complete mitochondrial genome sequence of Xenocypris davidi (Bleeker). Mitochondrial DNA. 25:374–376.
- Sung W, Peiqi Y, Yiyu C. 1998. Endangered species scientific commission. China red data book of endangered. Animals: Pisces Science Press.
- Wang X, Huang Y, Liu N, Yang J, Lei F. 2015. Seven complete mitochondrial genome sequences of bushtits (Passeriformes, Aegithalidae, Aegithalos): the evolution pattern in duplicated control regions. Mitochondrial DNA. 26:350–356.
- Wang S, Xie Y. 2004. China species red list, vol. 1. Red List. Higher Education. Beijing. p 159.