Abstract
In the present study, we presented the complete mitochondrial genome of Aspergillus sp. The complete mitochondrial genome of Aspergillus sp. was composed of circular DNA molecules, with a total length of 31,374 bp. The base composition of this mitochondrial genome is as follows: A (36.25%), T (38.09%), G (14.55%), and C (11.10%). The mitochondrial genome contains 18 protein-coding genes, 2 ribosomal RNA genes (rRNA), and 31 transfer RNA genes. The taxonomic status of the Aspergillus sp. mitochondrial genome exhibits a closest relationship with Aspergillus ustus and A. flavus.
Aspergillus is a group of filamentous fungi consisting of a few hundred species (Geiser Citation2009), which is found everywhere worldwide (Sugui et al. Citation2014). Aspergillus spp. spread as a contaminant of edible fungi by air, water, and plant debris, which commonly cause allergic and occasionally life-threatening infections in humans (Enoch et al. Citation2006). Some Aspergillus species affect humans, animals, and birds, which can develop aspergillosis (Gautier et al. Citation2016; Paulussen et al. Citation2017). Many plant diseases and food spoilage may also be due to Aspergillus infection (Amaike and Keller Citation2011). The mitochondrial genome of Aspergillus sp. reported here will promote further understanding of the population genetics, evolution, and taxonomy of this important fungal group.
The specimen (Aspergillus sp.) was isolated from the contaminated cultivation material of Auricularia polytricha in Chengdu, Sichuan, China (104.42 E; 30.88 N) and was stored in Sichuan Academy of Agricultural Sciences (No. As_1). The sample was initially identified as Aspergillus spp. based on phenotypic characteristics (Varga et al. Citation2008). Fungal DNA Kit D3390-00 (Omega Bio-Tek, Norcross, GA) was used to extract the total genomic DNA of Aspergillus sp. Then we purified the genomic DNA through a Gel Extraction Kit (Omega Bio-Tek). The purified genomic DNA was stored in the sequencing company (BGI Tech, Shenzhen, China). Sequencing libraries were constructed with purified DNA following the instructions of NEBNext® Ultra™ II DNA Library Prep Kit (NEB, Beijing, China). Whole genomic sequencing was performed by the Illumina HiSeq 2500 Platform (Illumina, SanDiego, CA). Multiple steps were used for quality control and de novo assembly of the mitogenome according to Qiang et al. (Citation2018). The SPAdes 3.9.0 software (Bankevich et al. Citation2012) was used to assemble the mitochondrial genome of Aspergillus sp. Gaps among contigs were filled using MITObim V1.9 (Hahn et al. Citation2013). The complete mitochondrial genome was annotated using the MFannot tool (Valach et al. Citation2014), combined with manual corrections. Transfer RNA (tRNA) genes were predicted using tRNAscan-SE v1.3.1 (Lowe and Chan Citation2016).
The total length of Aspergillus sp. mitogenome is 31,374 bp. This mitochondrial genome was submitted to GenBank database under accession No. MK038875. The circular mitogenome contains 18 protein-coding genes, 2 ribosomal RNA genes (rns and rnl), and 25 tRNA genes. The base composition of the genome is as follows: A (36.25%), T (38.09%), G (14.55%), and C (11.10%).
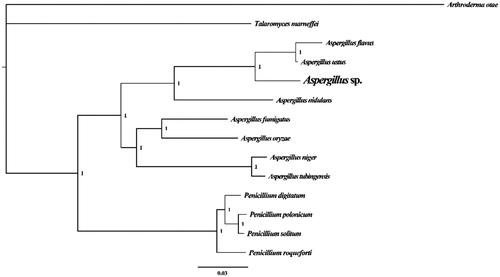
To validate the phylogenetic position of Aspergillus sp., we construct the phylogenetic trees of 14 closely related species based on the nucleotide sequences of the 14 core PCGs, in addition to the rns and rnl mitochondrial genes. Bayesian inference phylogenetic methods were used to construct phylogenetic trees using the combined gene datasets with MrBayes v3.2.6 (Ronquist et al. Citation2012). Bayesian posterior probabilities were calculated to assess node support. As shown in the phylogenetic tree (), the taxonomic status of the Aspergillus sp. based on combined mitochondrial gene dataset exhibits a closest relationship with A. ustus and A. flavus (Joardar et al. Citation2012).
Figure 1. Molecular phylogenies of 13 species based on Bayesian inference analysis of the combined mitochondrial gene set (14 core protein-coding genes + 2 rRNA genes). Node support values are Bayesian posterior probabilities. Mitogenome accession numbers used in this phylogeny analysis: Aspergillus flavus (NC_026920), Aspergillus fumigatus (NC_017016), Aspergillus nidulans (NC_017896), Aspergillus niger (NC_007445), Aspergillus oryzae (NC_018100), Aspergillus tubingensis (NC_007597), Aspergillus ustus (NC_025570), Penicillium digitatum (NC_015080), Penicillium polonicum (NC_030172), Penicillium roqueforti (NC_027416), Penicillium solitum (NC_016187), Talaromyces marneffei (NC_005256), Arthroderma otae (NC_012832).
Disclosure statement
The authors have declared that no competing interests exist.
Additional information
Funding
References
- Amaike S, Keller NP. 2011. Aspergillus flavus. Annu Rev Phytopathol. 49:107–133.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Enoch DA, Ludlam HA, Brown NM. 2006. Invasive fungal infections: a review of epidemiology and management options. J Med Microbiol. 55:809–818.
- Gautier M, Normand AC, Ranque S. 2016. Previously unknown species of Aspergillus. Clin Microbiol Infect. 22:662–669.
- Geiser DM. 2009. Sexual structures in Aspergillus: morphology, importance and genomics. Med Mycol. 47:S21–S26.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Joardar V, Abrams NF, Hostetler J, Paukstelis PJ, Pakala S, Pakala SB, Zafar N, Abolude OO, Payne G, Andrianopoulos A, et al. 2012. Sequencing of mitochondrial genomes of nine Aspergillus and Penicillium species identifies mobile introns and accessory genes as main sources of genome size variability. BMC Genomics. 13:698.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Paulussen C, Hallsworth JE, Álvarez-Pérez S, Nierman WC, Hamill PG, Blain D, Rediers H, Lievens B. 2017. Ecology of aspergillosis: insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb Biotechnol. 10:296–322.
- Qiang L, Mei Y, Cheng C, Chuan X, Xin J, Zhigang P, Wenli H. 2018. Characterization and phylogenetic analysis of the complete mitochondrial genome of the medicinal fungus Laetiporus sulphureus. Sci Rep. 8:9104.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Sugui JA, Kwon-Chung KJ, Juvvadi PR, Latgé JP, Steinbach WJ. 2014. Aspergillus fumigatus and related species. Cold Spring Harb Perspect Med. 5:a019786.
- Valach M, Burger G, Gray MW, Lang BF. 2014. Widespread occurrence of organelle genome-encoded 5S rRNAs including permuted molecules. Nucleic Acids Res. 42:13764–13777.
- Varga J, Houbraken J, Van Der Lee HA, Verweij PE, Samson RA. 2008. Aspergillus calidoustus sp. nov., causative agent of human infections previously assigned to Aspergillus ustus. Eukaryotic Cell. 7:630–638.

