Abstract
The complete mitogenome of Asterorhombus intermedius is 16,886 bp in length, containing 37 genes, among them, ND6 and eight tRNA genes are encoded by L-strand, and other genes by H-strand, which are the same as those of typical mitogenome in fishes. A striking finding is a novel genomic-scale gene rearrangement related nine genes, including ND6 and six tRNA encoded by the L-strand grouped to a cluster of Gln-Ala-Cys-Tyr-Ser1-ND6-Glu. Also, the genes of this cluster with identical transcriptional polarities maintained the original gene order. In addition, the order of 12S-Val-16S-Leu1 fragment rearranged to 12S-16S-Val-Leu1, and that of Ser1-Asp-COII fragment rearranged to Ser1-Asp-ND6.
Asterorhombus intermedius (Bleeker 1865) belongs to the family Bothidae of Pleuronectiformes (Amaoka Citation1969). The mitochondrial genome of A. intermedius were obtained (GenBank accession number: MK256952) based on one sample collected from Sanya City, Hainan, China (N18°16′33.87″, E109°43′33.24″). This specimen was stored in the Marine Biodiversity Collection of South China Sea, Chinese Academy of Sciences, under the voucher number SCF2015121552.
The mitochondrial genome of A. intermedius is 16,886 bp in length, including 13 protein-coding genes, two ribosomal RNA (rRNA), 22 transfer RNA (tRNA), and a control region (CR). Among these genes, 28 genes are encoded on the H-strand, others including ND6 and eight tRNA genes on the L-strand. The overall base composition is 27.6, 30.0, 16.5, and 25.9% for A, C, G, T, respectively, with a slight bias on AT content (53.5%).
The size of 13 protein-coding genes ranges from 168 bp (ATP8) to 1818 bp (ND5), that of 12S and 16S rDNA are 944 bp and 1653 bp, respectively and that of CR is 1042 bp. The size of 22 tRNA genes ranges from 65 bp (tRNA-Cys) to 73 bp (tRNA-Leu2), which were identified using tRNAscan-SE 2.0 (Lowe and Chan Citation2016).
A striking finding of this study is a novel genomic-scale gene rearrangement related nine genes, which was not similar to both of the typical fishes and other bothid species (Wang et al. Citation2015; Gong et al. Citation2016; Li et al. Citation2015). In the mitogenome of this species, the ND6 and six tRNA encoded by the L-strand grouped to a seven-gene cluster of Gln-Ala-Cys-Tyr-Ser1-ND6-Glu at the position between tRNA-T and tRNA-P. Besides, the genes of this cluster with identical transcriptional polarities maintained the original gene order. Also, the tRNA-Asp translocated from the position between tRNA-Ser1 and COII to the position between tRNA-Ser2 and ND6, which formed the eight-gene order of Gln-Ala-Cys-Tyr-Ser1-Asp-ND6-Glu. In addition, the order of 12S-Val-16S-Leu1 fragment rearranged to 12S-16S-Val-Leu1.
This mitogenome existed in 20 intergenic spacers with total 313 bp in length. Comparing with that of non-rearranged species, 10 abnormal intergenic regions with total 282 bp in length were found, among them, the longest is 98 bp and the shortest is 4 bp.
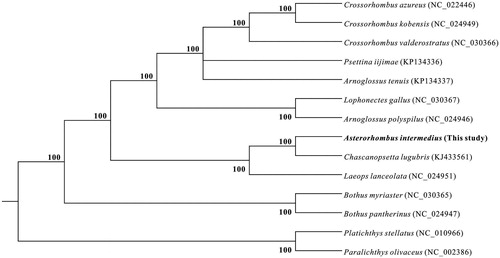
The phylogenetic tree was constructed based on the mitogenome of A. intermedius and other 11 bothid species with Paralichthys olivaceus and Platichthys stellatus as outgroup by using Bayesian inference in MrBayes 3.2 (Ronquist and Huelsenbeck Citation2003). The mitogenome sequences of 11 species and 3 outgroups were retrieved from GenBank. In the phylogenetic tree (), three species of Crossorhombus and two species of Bothus formed two clades, respectively, both with 100% posterior probability. However, two species of Arnoglossus were not clustered together. A. intermedius and Chascanopsetta lugubris formed a clade with 100% posterior probability, which means A. intermedius has a relatively closer phylogenetic relationship to C. lugubris, compared with other 10 bothid species.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Amaoka, K. 1969. Studies on the sinistral flounders found in the waters around Japan: taxonomy, anatomy and phylogeny. J Natl Fish Univ. 18:65–340.
- Gong L, Shi W, Yang M, Li DH, Kong XY. 2016. Novel gene arrangement in the mitochondrial genome of Bothus myriaster (Pleuronectiformes: Bothidae): evidence for the dimer-mitogenome and non-random loss model. Mitochondrial DNA A DNA Mapp Seq Anal. 27:3089–3092.
- Li DH, Shi W, Munroe TA, Gong L, Kong XY. 2015. Concerted evolution of duplicate control regions in the mitochondria of species of the flatfish family Bothidae (Teleostei: Pleuronectiformes). PLoS One. 10:e0134580.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–57.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Wang SY, Shi W, Wang ZM, Gong L, Kong XY. 2015. The complete mitochondrial genome sequence of Aesopia cornuta (Pleuronectiformes: Soleidae). Mitochondrial DNA. 26:114–115.