Abstract
Cyprinus carpio triploid is induced by temperature shock technology. It has been extensively cultivated and got great economic value in China. The complete mitochondrial genome of C. carpio triploid is the 16,580 bp length and contains 37 genes (13 protein-coding genes (PGCs), two ribosomal RNA, 22 transfer RNA genes, and one control region). The overall base composition of the heavy strand is that the 31.92% for A, the 27.53% for C, the 15.73% for G, and the 24.83% for T. The percentage of G + C content is 43.26% and A + T content is 56.74%. The result of phylogenetic analysis indicated that C. carpio triploid was clustered in genus Cyprinus as expected. The complete mitogenome may provide important date set for genetic status and the evolutionary study of genetic mechanism of C. carpio.
Cyprinus carpio triploid (Cypriniformes, Cyprinidae, Cyprinus.) is induced by polyploidization technology. Polyploid species possess a greater capability to withstand ecological variation because their genes have more metabolic flexibility (Otto and Whitton 2000). Cyprinus carpio triploid has been extensively cultivated and got great economic value in China (Xu et al. Citation2014).
In this work, C. carpio triploid collected from Heilongjiang Fisheries Research Institute Kuandian aquaculture experimental station was used for the complete mitochondrial DNA sequencing. The total genomic DNA was extracted from alcohol-preserved fin using the traditional phenol-chloroform method (Taggart et al. Citation1992). Total genomic DNA was extracted from tail tip using the Ezup pillar genomic DNA extraction kit (Sangon Biotech, Shanghai, China). A total of 13 pairs of primers, designed by based on the mitochondrial genome of C. carpio (X61010.1) (Chang et al. Citation1994), were used to amplify the complete mitochondrial genome. The PCR products were sequenced using Sanger method and further assembled into a sequence with CAP3 (Huang and Madan Citation1999).
Similar to the typical vertebrate mitogenomes, the complete mitochondrial DNA of C. carpio triploid was a circular molecule with 16,580 bp in length (GenBank accession number MK134852). The mitochondrial genome of comprised of 37 genes including 13 protein-coding genes, two ribosomal RNA, 22 transfer RNA genes, and one control region. The major non-coding sequence of mitochondrial genome was the control region (D-loop), which was located between the tRNA Pro and tRNA Phe genes with 926 bp in length. The overall base composition of the heavy strand is 31.92% for A, 27.53% for C, 15.73% for G, and the 24.83% for T. The percentage of GC content is 43.26% and AT content is 56.74%.
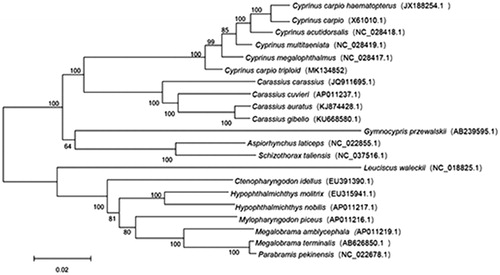
We constructed a phylogenetic tree including C. carpio triploid and the other 20 fish. Phylogenetic relationships obtained with the maximum likelihood approach were identical to those of the Bayesian analysis (Posada and Buckley Citation2004; Wang et al. Citation2015). As shown in the phylogenetic tree (), C. carpio triploid sequence was clustered in genus Cyprinus, including C. carpio haematopterus, Cyprinus multitaeniata, Cyprinus acutidorsalis, Cyprinus megalophthalmus, and Cyprinus multitaeniata. The genus Cyprinu has a close relationship with the genus Carassius. The specimen was stored in the aquatic species library of Northeast Agricultural University.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chang YS, Huang FL, Lo TB. 1994. The complete nucleotide sequence and gene organization of carp (Cyprinus carpio) mitochondrial genome. J Mol Evol. 38:138–155.
- Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868–877.
- Otto SP, Whitton J. 2000. Polyploid incidence and evolution. Annu Rev Genet. 34:401–437.
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 53:793–808.
- Taggart JB, Hynes RA, Prodouhl PA, Ferguson A. 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. Journal of Fish Biology. 40:963–965.
- Wang X, Huang Y, Liu N, Yang J, Lei F. 2015. Seven complete mitochondrial genome sequences of bushtits (Passeriformes, Aegithalidae, Aegithalos): the evolution pattern in duplicated control regions. Mitochondrial DNA. 26:350–356.
- Xu P, Zhang X, Wang X, Li J, Liu G, Kuang Y, Xu J, Zheng X, Ren L, Wang G, et al. 2014. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat Genet. 46:1212–1219.