Abstract
The complete mitochondrial genome of Scatella tenuicosta has been reported in this study. This is the first sequenced complete mitogenome of the genus Scatella. Totally, 37 genes presented in the 15,968 bp circular genome consisting of 22 transfer RNA genes, 13 protein-coding genes, 2 ribosomal RNA genes, and a non-coding A + T rich-region. The phylogenetic tree was constructed based on the 13 concatenated PCGs using Bayesian inference method and showed that S. tenuicosta formed a monophyletic clade, sister to Archisepsis discolor.
Ephydroidea is divided into five families comprising of Curtonotidae, Drosophilidae, Camillidae, Diastatidae, and Ephydridae (McAlpine Citation1989), with 16 genera in Ephydrinae. Scatella Robineau-Desvoidy, which belongs to the subfamily Ephydrinae, is one of the most species-rich genera of shore flies in the world with 135 described species (Mathis and Zatwarnicki Citation1995). For the moment, just some partial sequences have been deposited in the GenBank database, but none of the mitochondrial genome of Scatella sp. was available. In this study, we sequenced the complete mitochondrial genome of S. tenuicosta (GenBank accession number: MK353462) and analyzed its phylogeny.
In this paper, the samples of S. tenuicosta, which we could mine from the stems and roots of seedling of the family Cucurbitaceae, were collected from Huangpi District (30°42′35″N, 114°28′11″E), Wuhan city, Hubei Province, China. The specimens now are deposited in Wuhan Academy of Agricultural Sciences. High-throughput sequencing method (Tian et al. Citation2019) was employed to determine the complete mitogenome of S. tenuicosta with further bioinformatic analysis. The total mitogenome of S. tenuicosta has a length of 15,968 bp. It shows a conserved arrangement pattern, including 13 PCGs, 22 tRNA genes, 2 rRNA genes, and a large non-coding A + T rich-region. In this study, standard start codons ATN were commonly used by most PCGs and NAD1 started with TTG (Wolstenholme Citation1992), except for Cox1 which used TCG (Li et al. Citation2016). COX1, COX2, NAD5, and NAD1 finished by T–, and other PCGs ended with TAA. Among 37 genes, 23 genes were oriented on the heavy (H) strand, others on the light (L) strand. All tRNAs in mitochondrial genomes of S. tenuicosta arranged from 63 bp to 72 bp. The control region is 1054 bp in length with A + T contents of 89.9% and located between the 12s rRNA and tRNAIle-tRNAGln-tRNAMet cluster. The overall base compositions of A, C, T, and G are 40.04, 12.73, 9.29, and 37.94%, respectively. The nucleotide compositions of the mitochondrial genomes are remarkably AT-biased (78.0%).
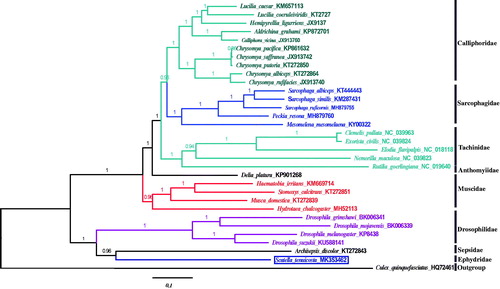
To determine the phylogenetic position of S. tenuicosta, phylogenetic analysis was based on the nucleotide sequences of the 13 PCGs of 32 closely related species and performed using MrBayes v3.2.6 (Ronquist et al. Citation2012) with GTR + I+invgamma substitution model, as selected by ModelTest3.7. As shown in the phylogenetic tree (), S. tenuicosta formed a monophyletic group separately and it was closest to A. discolor in the family Sepsidae with high-nodal support (BI posterior probabilities [PP] = 0.96), then clustered with the Drosophilidae species. In addition, the species in the family Calliphoridae, Sarcophagidae, Tachinidae, Anthomyiidae, and Muscidae formed another phylogroup. The complete mitochondrial data would benefit to species delimitation, the taxonomic status and phylogenetic analysis of ephydrid in family Ephydridae.
Figure 1. Phylogenetic position of S. tenuicosta based on a comparison with 13 concatenated PCGs of 31 other species in Diptera using Bayesian tree estimate methods. Numbers on branches indicated posterior probablility. Culex quinquefasciatus was used as outgroup. GeneBank accession numbers of each species were listed in the tree after the scientific name.
Acknowledgments
We express our sincere thanks to Professor Ding Yang and Dr. Liang Wang from the Department of Entomology, China Agricultural University, Beijing, P.R. China, for the morphological identification of S. tenuicosta. We thank Dr. Mang Shi from the University of Sydney, Sydney, New South Wales, Australia, for the assistance with high_throughput data analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Li X, Wang Y, Su S, Yang D. 2016. The complete mitochondrial genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1435–1436.
- Mathis WN, Zatwarnicki T. 1995. A world catalog of the shore flies (Diptera: Ephydridae). Memoirs on Entomology, International. 4:1–423.
- McAlpine JF 1989. Phylogeny and classification of the muscomorpha introduction. In McAlpine JF WoodDM editors Manual of nearctic diptera. Vol. 3. Agriculture Canada Monograph, 28; 1333p–1581; 1397–1502.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Tian JH, Ge MH, Xu HB, Wu TP, Yu B, Lei CL. 2019. The complete mitochondrial genome and phylogenetic analysis of Haemaphysalis hystricis (Parasitiformes: Ixodidae). Mitochondr DNA B. 4:1049–1050.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.
- Zatwarnicki T. 1992. A new classification of Ephydridae based on phylogenetic reconstruction (Diptera Cyclorrhapha). Genus. 3:65–119.

