Abstract
The dance fly Empis separata belongs to the subfamily Empidinae of Empididae. The mitogenome (GenBank accession number: MK993569) of E. separata was sequenced, the new representative of the mitogenome of the subfamily. The complete mitogenome is 14,961 bp totally, consisting of 13 protein-coding genes, two rRNAs, and 22 transfer RNAs. All genes have the similar locations and strands with that of other published species of Empididae. The nucleotide composition biases toward A and T, which together made up 76.3% of the entirety. Bayesian inference analysis strongly supported the monophyly of Empidoidea, Empididae, and Dolichopodidae. This result also suggested that Empidinae is the sister group to the clade of Trichopezinae.
Introduction
Empididae is one of the largest families in Diptera with over 5,000 described species from the world (Yang et al. Citation2007). They capture aphids, psyllids, and coccids of Hemiptera, but also other true flies such as mosquitos, blackflies, and so on. They are widely used as a biological indicator of evaluating the quality of environment and biodiversity (Yang and Yang Citation2004).
The specimens of E. separata used for this study were collected in Zhouzhi County of Shaanxi by Jinjin Ning and identified by Ding Yang. Specimens are deposited in the Entomological Museum of China Agricultural University (CAU) with the accession number: CAU201583. The total genomic DNA was extracted from the whole body (except head) of the specimen using the QIAamp DNA Blood Mini Kit (Qiagen, Germany) and stored at −20 °C until needed. The mitogenome was amplified and sequenced as described in our previous study (Wang et al. Citation2016). The nearly complete mitogenome of E. separata is 14,961 bp (GenBank accession number: MK993569). It encoded 13 PCGs, 22 tRNA genes, two rRNA genes, and the control region could not be sequenced entirely in this study and was similar with related reports before (Li et al. Citation2016; Wang, Ding, et al. Citation2016; Wang, Wang, et al. Citation2016; Li et al. Citation2017; Zhou et al. Citation2017; Qilemoge et al. Citation2018; Gao et al. Citation2018). All genes have the similar locations and strands with that of other published Empididae species. The nucleotide composition of the mitogenome was biased toward A and T, with 76.3% of A + T content (A = 38.5%, T = 37.8%, C = 14.1%, and G = 9.5%). The A + T content of PCGs, tRNAs, and rRNAs is 75.0%, 77.8%, and 81.5%, respectively. The total length of all 13 PCGs of E. separata is 11,301 bp. Five PCGs (NAD2, ATP8, NAD3, NAD5, and NAD6) initiated with ATT codons, and six PCGs (COII, COIII, ATP6, NAD4, NAD4L, and CYTB) initiated with ATG codons, NAD1 initiated with TTG as a start codon, and CO1 initiated with TCG as a start codon. Thirteen PCGs used the typical termination codons TAA in E. separata.
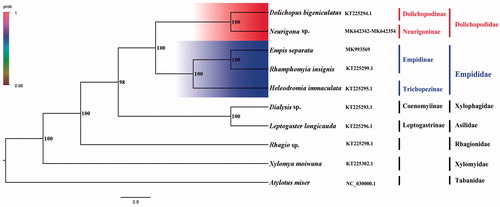
Phylogenetic analysis was performed based on the nucleotide sequences of 13 PCGs from 10 Diptera species. Bayesian (BI) analysis generated the phylogenetic tree topologies based on the PCGs matrices (). According to the phylogenetic result, the monophyly of Empidoidea was strongly supported. The monophyletic Dolichopodidae that contains Dolichopodinae and Neurigoninae was assigned to the sister group to the clade of Empididae that consists of Empidinae and Trichopezinae in this study. For the phylogeny within Empidoidea, the Empidinae is the sister to the clade of Trichopezinae, and Dolichopodinae is the sister to the clade of Neurigoninae. This result show that Dolichopodidae is the sister group to Empididae, which is consistent with the phylogenetic result of the previous research (Wang et al. Citation2016). The mitogenome of E. separata could provide the important information for the further studies of Empidoidea phylogeny.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Gao S, Zhou J, Cao Y, Zhang J, Yang D. 2018. The mitochondrial genome of Trichoclinocera yixianensis (Diptera: Empididae). Mitochondrial DNA B. 3:452–453.
- Li X, Ding S, Hou P, Liu X, Zhang C, Yang D. 2017. Mitochondrial genome analysis of Ectophasia rotundiventris (Diptera: Tachinidae). Mitochondrial DNA B. 2:457–458.
- Li X, Wang Y, Su S, Yang D. 2016. The complete mitochondrial genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1435–1436.
- Qilemoge Gao S, Tang C, Wang N, Yang D. 2018. The mitochondrial genome of Diostracus lamellatus (Diptera: Dolichopodidae). Mitochondrial DNA B. 3:976–347.
- Wang K, Ding S, Yang D. 2016. The complete mitochondrial genome of a stonefly species, Kamimuria chungnanshana Wu, 1948 (Plecoptera: Perlidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:3810–3811.
- Wang K, Li X, Ding S, Wang N, Mao M, Wang M, Yang D. 2016. The complete mitochondrial genome of the Atylotus miser (Diptera: Tabanomorpha: Tabanidae), with mitochondrial genome phylogeny of lower Brachycera (Orthorrhapha). Gene. 586:184–196.
- Wang K, Wang Y, Yang D. 2016. The complete mitochondrial genome of a stonefly species, Togoperla sp. (Plecoptera: Perlidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1703–1704.
- Yang D, Yang C. 2004. Fauna Sinica Insecta. Vol. 34. Diptera Empididae. Beijing: Science Press.
- Yang D, Zhang K, Yao G, Zhang J. 2007. World Catalog of Empididae (Insecta: Diptera). Beijing: China Agricultural University Press.
- Zhou Q, Ding S, Li X, Zhang T, Yang D. 2017. Complete mitochondrial genome of Allognosta vagans (Diptera, Stratiomyidae). Mitochondrial DNA B. 2:461–462.